Fig. 1. Boule circRNAs are conserved and required for male fecundity under heat stress in fruit flies.
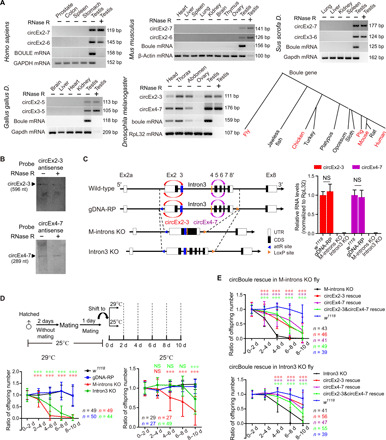
(A) RT-PCR [with or without ribonuclease (RNase) R treatment] of Boule circRNAs in human, mouse, pig, chicken, and fly. A phylogenetic diagram of Boule homologs in animals (animals examined for circBoule labeled in red). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; bp, base pairs. (B) Northern blot of circEx2-3 and circEx4-7 from wild-type (w1118) testes. nt, nucleotide. (C) Left: Scheme for generation of boule genomic DNA replacement (gDNA-RP), knockout (KO) of boule middle introns (introns 2, 3, 4, 5, 6, and 7; M-introns KO), and KO of boule intron 3 (Intron3 KO) flies. Right: Levels of circBoule RNAs in testes of 2-day-old w1118, gDNA-RP, M-introns KO, and Intron3 KO flies reared at 25°C. Ribosomal protein L32 (RpL32) mRNA was used as endogenous control. UTR, untranslated region. (D) Ratio of offspring numbers (normalized to progeny from 0 to 2 days) in w1118, gDNA-RP, M-introns KO, and Intron3 KO male flies mating at 29° or 25°C. Scheme of experimental setup is shown. Details of shifting male flies from 25° to 29°C are described in Materials and Methods. There was no significant difference in fertility between w1118 and gDNA-RP male flies at 29° or 25°C. Red or green asterisks and NS (not significant) indicate statistical analyses between gDNA-RP and M-introns KO or Intron3 KO flies. (E) Ratio of offspring numbers in circEx2-3 rescue, circEx4-7 rescue, or circEx2-3&circEx4-7 rescue male flies (29°C). CircEx2-3 rescue, circEx4-7 rescue, or circEx2-3&circEx4-7 rescue is overexpression of boule circEx2-3, circEx4-7, or circEx2-3&circEx4-7 in M-introns KO or Intron3 KO flies by β2-tubulin promoter. Red, purple, or green asterisks indicate statistical analyses of fertility between circEx2-3, circEx4-7, or circEx2-3&circEx4-7 rescue and the corresponding control (M-introns KO or Intron3 KO). In (D) and (E), n = number of male flies tested. Data are shown as means ± SD. P values from unpaired Student’s t test, ***P < 0.001.
