Fig. 1. Crystal structure of LMTK3.
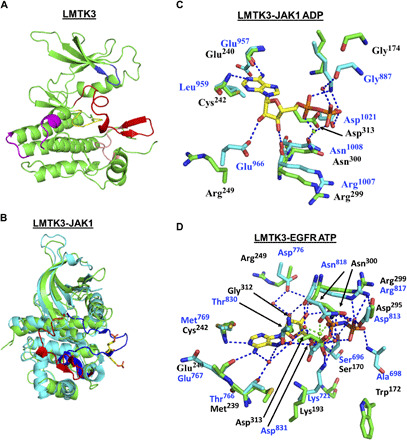
(A) Structure of the kinase domain of LMTK3 showing the main features [Protein Data Bank (PDB) 6SEQ]. Blue, the glycine-rich loop; red, activation loop; salmon, the P+1 loop. The activation loop and the P+1 loop constitute the activation segment. Yellow, the catalytic loop; magenta, the kinase insert region. (B) Superimposition of the kinase domains of LMTK3 (green) and JAK1 (cyan). The activation loop of LMTK3 is shown in red and adopts an inactive conformation. The activation loop of JAK1 is shown in blue and adopts the active state. The two JAK1 phosphotyrosine residues (Tyr1034 and Tyr1035) are shown as yellow sticks. (C) Superimposition of LMTK3 and JAK1 active site (PDB 5KHW) residues involved in ADP binding. Key residues that bind ADP in JAK1 are positioned slightly differently to those of LMTK3 because of the different catalytic states of the kinases. However, residues in JAK1 that interact with ADP are conserved within LMTK3, although two noticeable differences are seen (LMTK3 Arg249 and Cys242 replaces Glu966 and Leu959), which are unlikely to affect ATP binding. (D) Superimposition of LMTK3 and EGFR active site (PDB 5CNO) residues involved in ATP binding. A comparison with the ATP-bound form of EGFR also shows strong conservation among residues of EGFR that bind ATP and amino acid residues at equivalent positions within LMTK3 and residues that are not conserved (Ala698, Met769, and Cys773 for Trp172, Cys242, and Asp246 in LMTK3), form main-chain interactions with bound ATP. [Green, amino acid residues of LMTK3 with black residue numbers; cyan, amino acid residues of JAK1 (top) or EGFR (bottom) with blue residue numbers; red spheres, water molecules; blue dashes, hydrogen bonds; green sphere, magnesium ion; green dashes, bonds coordinating the magnesium ion.]
