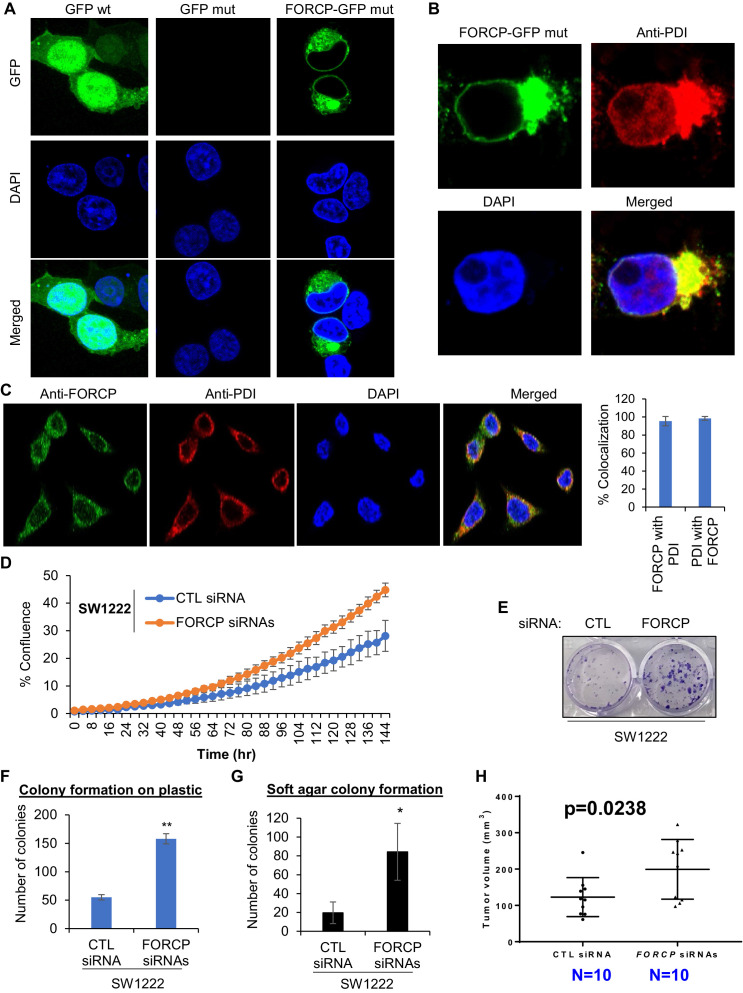
Figure 3. FORCP protein is localized to the endoplasmic reticulum and its knockdown leads to growth advantage.
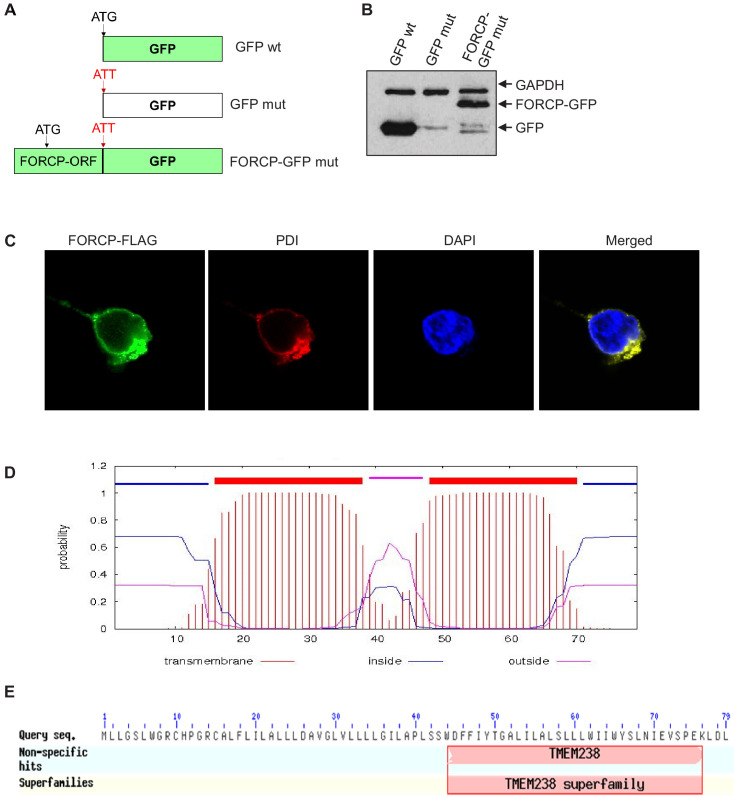
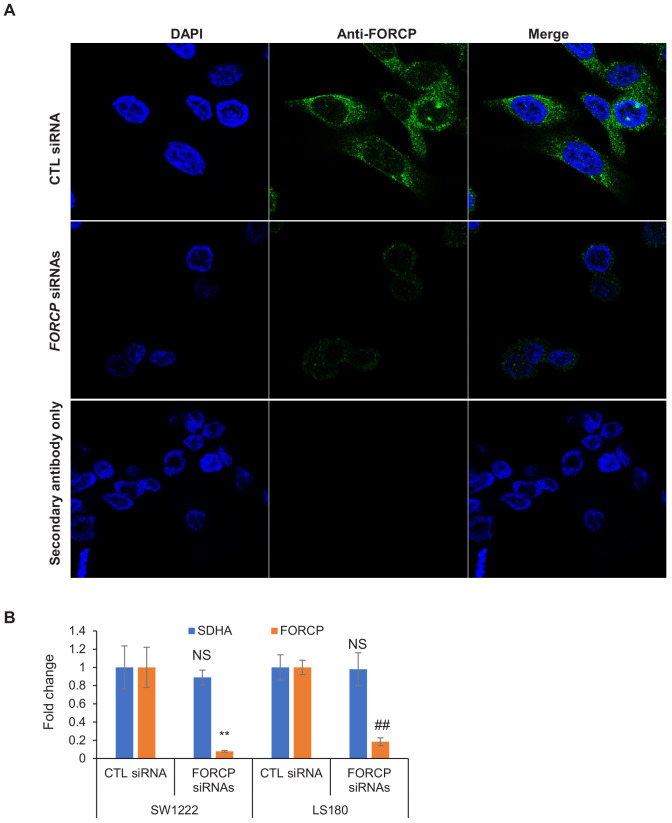
(A, B) Confocal microscopy was performed following transfection of 293 T cells with GFP wt, GFP with mutated ATG (GFP mut), or FORCP-GFP mut. Immunostaining for the ER marker PDI (B) shows colocalization of FORCP-GFP mut with PDI. DNA was counterstained with DAPI. (C) Confocal microscopy (left panel) following immunostaining of LS180 cells with anti-FORCP and anti-PDI antibodies shows colocalization of endogenous FORCP with the ER marker PDI. DNA was counterstained with DAPI. Colocalization of FORCP and PDI (right panel) was analyzed using ZEISS ZEN Desk microscope software from 100 individual cell images and converted to percentage co-localization. (D–H) The effect of FORCP knockdown in SW1222 cells on proliferation and tumorigenicity was assessed by Incucyte live cell proliferation assays (D), colony formation assays (E–G) and mouse xenograft experiments (H). Image from a representative colony formation on plastic experiment is shown in panel E and the data from three experiments is quantitated in panel F. For mouse xenograft experiments ‘N’ refers to the number of tumors 18 days after injecting the mice with SW1222 cells that were transfected for 48 hr with CTL or FORCP siRNAs. Error bars in panels D, F, and G represent SD from three experiments. *p< 0.05, **p< 0.005.