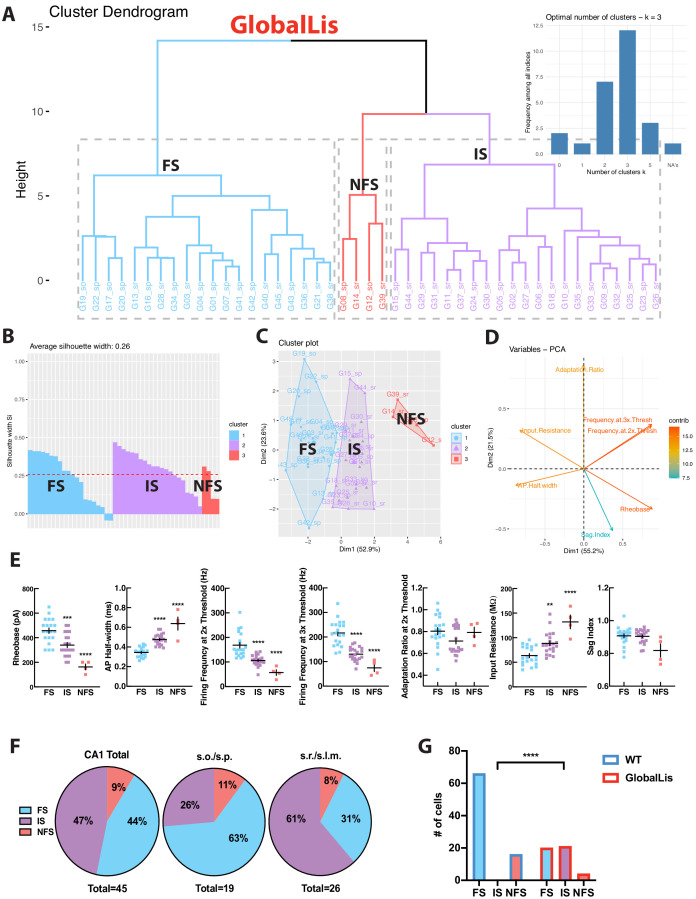
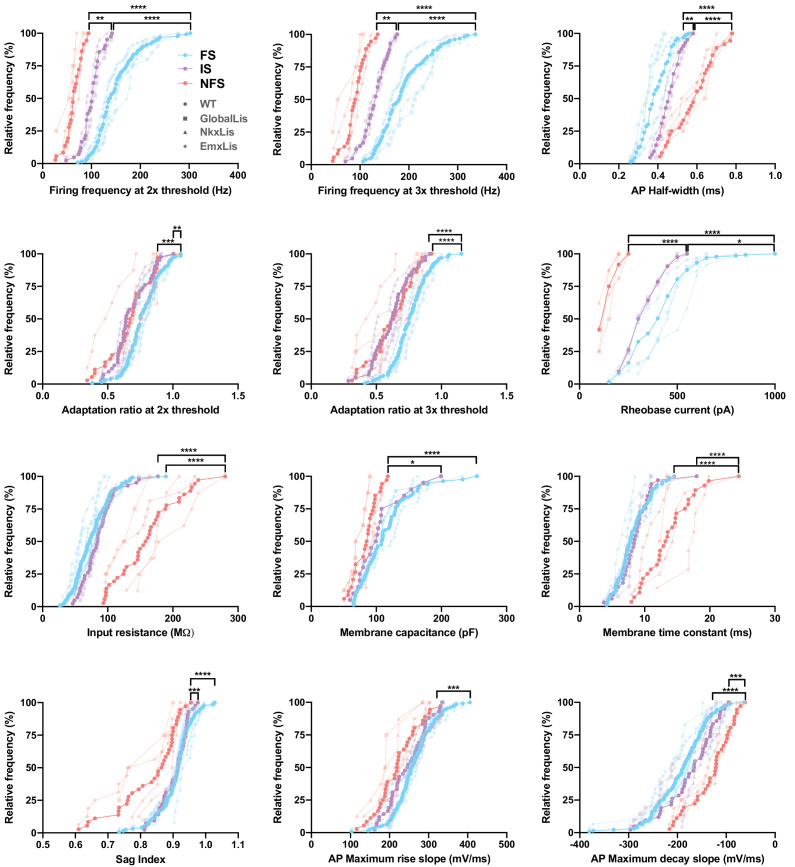
Figure 4. GlobalLis PV+INTs consist of three physiological subtypes: FS, IS, and NFS cells.
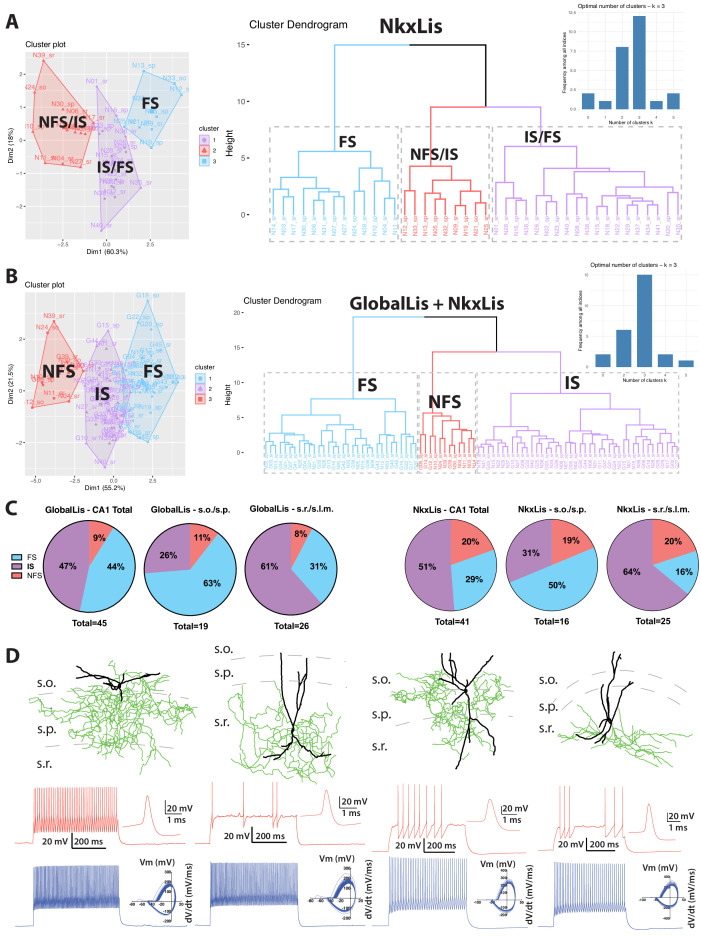
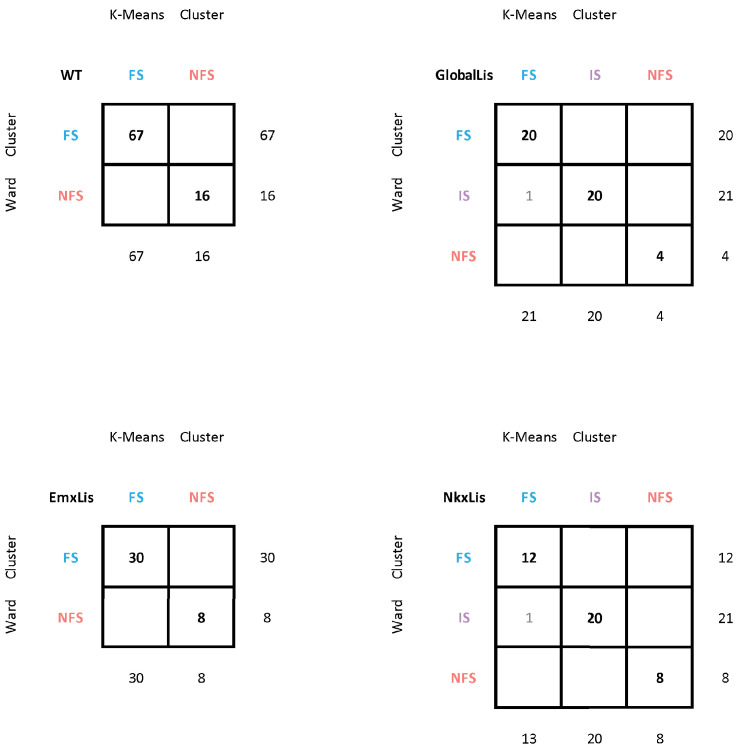
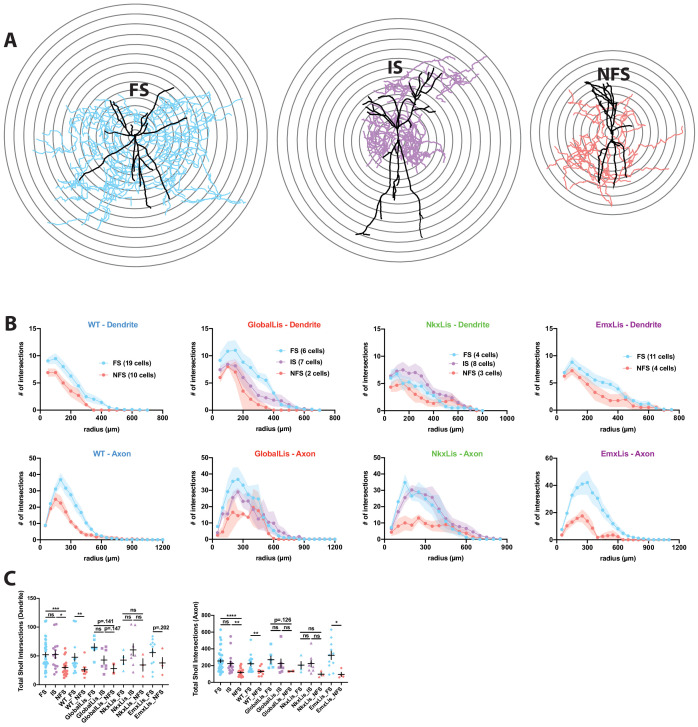
(A) Unbiased cluster analysis indicates emergence of an additional cluster in GlobalLis CA1, displayed in the dendrogram of 45 PV+INTs. Inset indicates optimal detection of three clusters (FS, IS, NFS). (B) Silhouette plot of the FS/IS/NFS clusters. (C) Cluster plot of the same 45 PV+INTs. (D) Contributions of each intrinsic physiological property used in the cluster analysis. (E) Plots displaying FS/IS/NFS cell intrinsic properties. For statistical analysis *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (F) Percentage of FS/IS/NFS cells in CA1 and in each sublayer. (G) Fisher exact test confirms differences in WT and GlobalLis composition of CA1 PV+INT subtypes.