FIG 3.
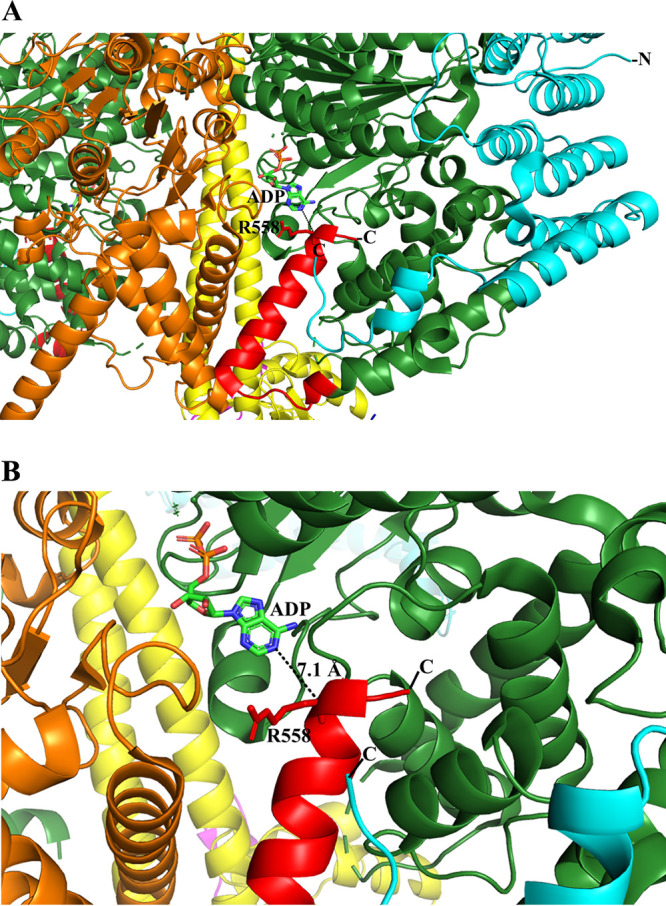
A proposed mechanism of ATP hydrolysis inhibition. (A) Part of the T. brucei F1-ATPase crystal structure (PDB ID 6F5D ) (22) and a further zoom to highlight the proximity of its extended subunit α C terminus and ADP. The T. brucei C-terminal residues 536 to 539 (red) form an α-helical turn, followed by a random region (540 to 544) and an α-helix (546 to 558) that come close to the ADP. A conformational alteration could bring R558 closer to ADP to generate a hydrogen bond with ADP, or one of the C-terminal residues, not resolved in the structure, could come in proximity to the nucleotide. We predict that the C-terminal residues 538 to 549 of mycobacterial subunit α may come in close proximity to the ADP to stabilize the inhibited state. Subunits α, β, and γ and the T. brucei-specific p18 are shown in green, orange, yellow, and cyan, respectively. The figure was generated via PyMOL (33).
