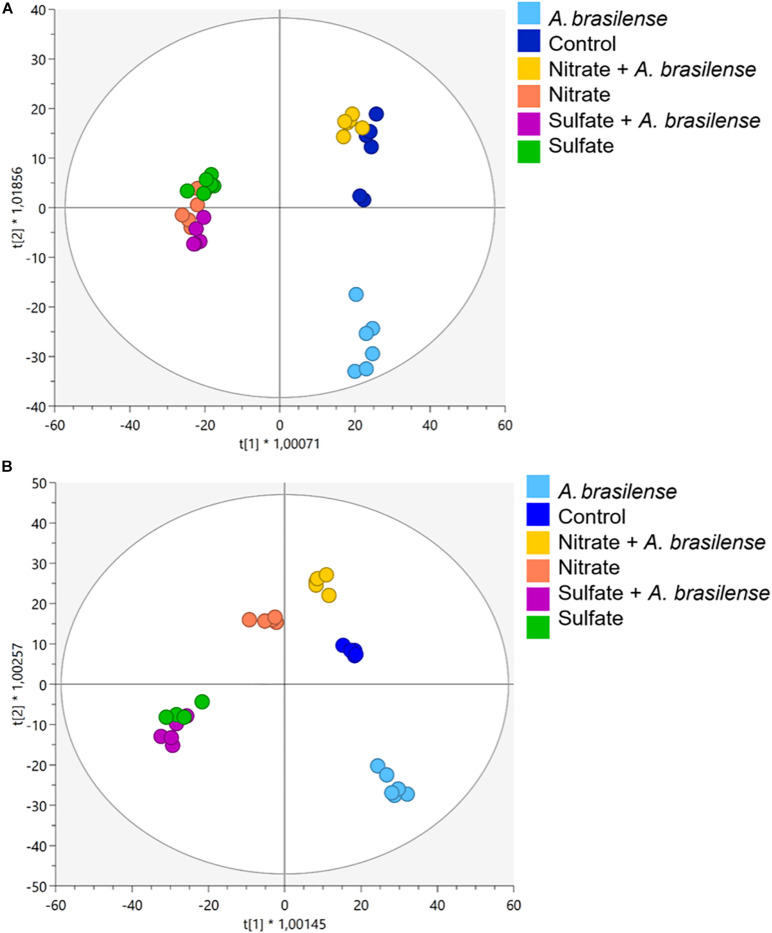
FIGURE 2.
Orthogonal projection to latent structures discriminant analysis (OPLS-DA) supervised modeling of metabolomic signatures in sweet basil plants of cv. (A) Genovese and (B) Red Rubin. Plants were grown in control hydroponic solution, in a NO3– or in a SO42– overfertilized nutrient solution, either non-inoculated or inoculated with A. brasilense. The metabolomic dataset produced through UHPLC-ESI/QTOF-MS was Pareto scaled and then used for the multivariate OPLS-DA modeling.