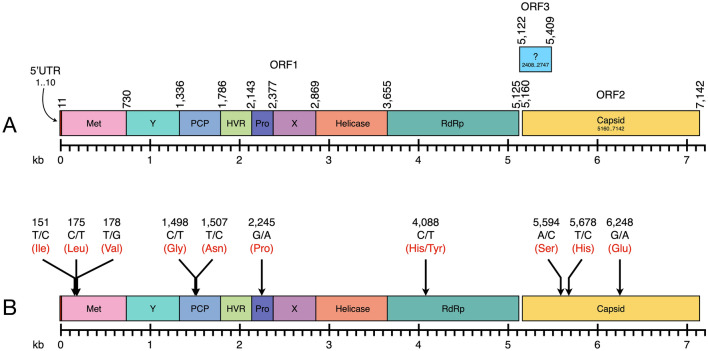
Fig. 2.
Genome organization of the Thai isolates of HEV (KY232312 and KY232313) and mutation events between those two genomes: the HEV genomes from serum and feces were identical in size and organization (a). The total length of the HEV genomes was 7142 bp, organized in three ORFs—1, 2, and 3. The ORF1 encoded a nonstructural polyprotein which contained at least eight regions, including a methyltransferase (Met), a Y domain (Y), a papain-like cysteine protease (PCP), a hypervariable region (HVR), a proline-rich region (Pro), a protein with an unknown function (X), a helicase (Helicase), and an RNA-dependent RNA polymerase (RdRp). The ORF2 encoded a single capsid gene (Capsid), and the ORF3 also carried a single protein with unknown function. The numbers on the genome diagram represent the boundary of each protein in the genome. Nucleotide substitution sites on the two genomes are shown by arrows and are labeled in black numbers (b). The labels at each arrow tail represent the nucleotide substitution site, pattern of nucleotide-substitution, and amino acid. The nucleotide in the HEV genome isolated from serum (KY232312), and feces (KY232313) are separated by a slash (/). The amino acids encoded from the mutation position are shown in red. Regarding the non-synonymous substitution, the amino acids in the different genomes are separated by a slash (/)