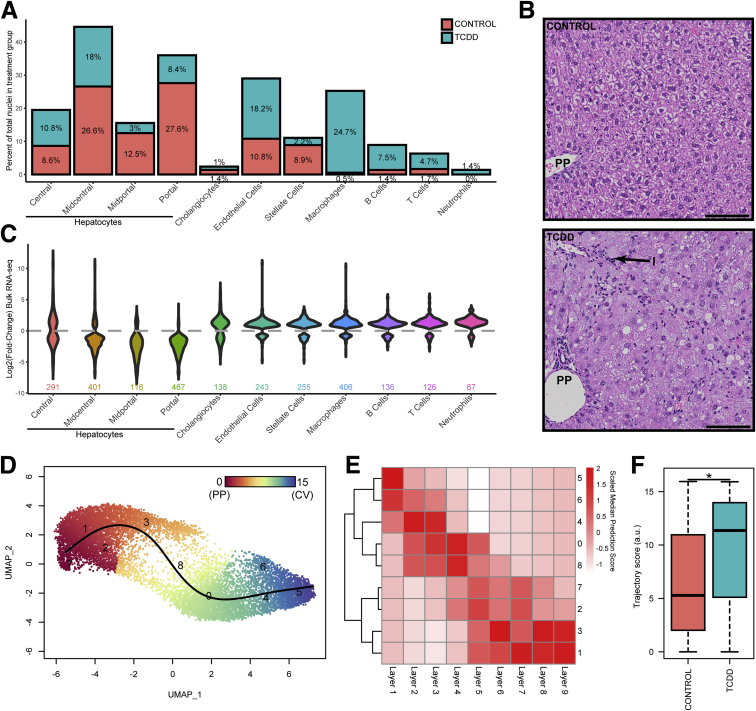
Figure 2.
Hepatic cell population shifts in response to TCDD. (A) Percentage of nuclei represented in cell type–specific clusters in control and treated samples. (B) Representative liver photomicrographs from control and TCDD-treated samples using the same study design33 showing periportal (PP) immune cell infiltration (I). Scale bar = 100 μm. (C) Violin plots of fold-change expression distribution for genes defining cell type clusters (adjusted P ≤ .05) in TCDD-elicited bulk RNA-seq dataset. (D) Reclustering and trajectory analysis of only hepatocyte nuclei from vehicle control and TCDD treatment groups using a resolution of 1.2 to identify 9 clusters in order to (E) compare snSeq gene expression with the 9 distinct spatially resolved hepatocyte layers defined Halpern et al.16 (F) Nuclei distribution along the trajectory determined in control and TCDD-treated samples. The asterisk indicates a significant difference in distribution with TCDD causing a shift in the hepatocyte nuclei population toward the central cluster/layer as determined by the Kolmogorov-Smirnov test (P ≤ .05).