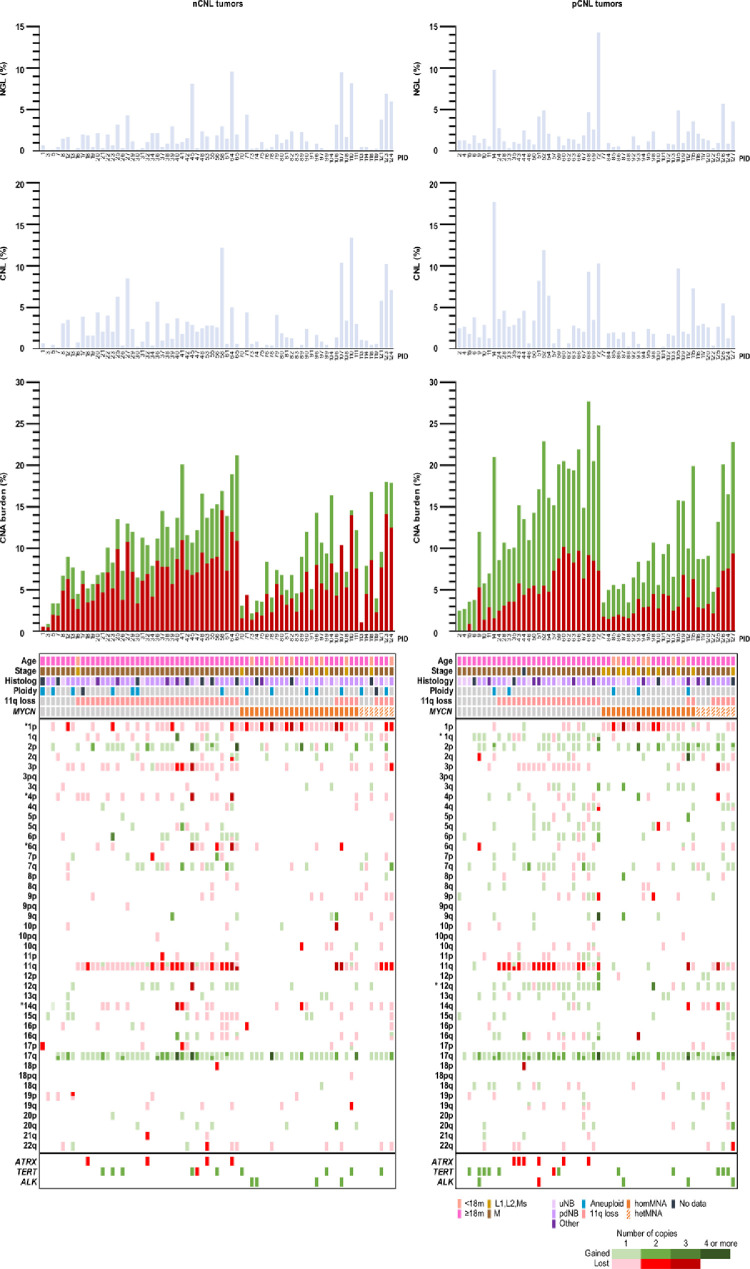
Figure 5.
Distribution of clinical and biological features according to copy number load genomic subtypes. The 127 HR-NB cases are classified according to CNL genomic subtype (left: nCNL tumors, 71 patients; right: pCNL tumors, 56 patients) and ranged by patterns of SCAs defined by MYCN status, from least to greatest number of SCAs to facilitate comparison of clinical and biological data. The SCAs in the different chromosomal regions are represented with a sliding scale from light to dark according to number of copies gained or lost with respect to the somy of the chromosome inferred from SNPa (green: gains, red: losses). When there are two SCAs in the same chromosome arm (p or q), they are different colored in the same region (bottom region in p arms and upper region in q arms are closer to centromere). For non-MNA tumors, loss of 1p, 4p, 6q and 14q is more frequent in nCNL tumors and gain of 1q and 12q is more frequent in pCNL tumors (marked with a star). Focal deletions in ATRX in non-MNA tumors and focal gains and losses in TERT and ALK are independent of the CNL genomic subtype. No difference was found in genomic instability indexes shown in the histograms nor in clinical and biological characteristics represented in the chart below. Patient identifiers (PID) are shown on the x-axes.