Figure 6.
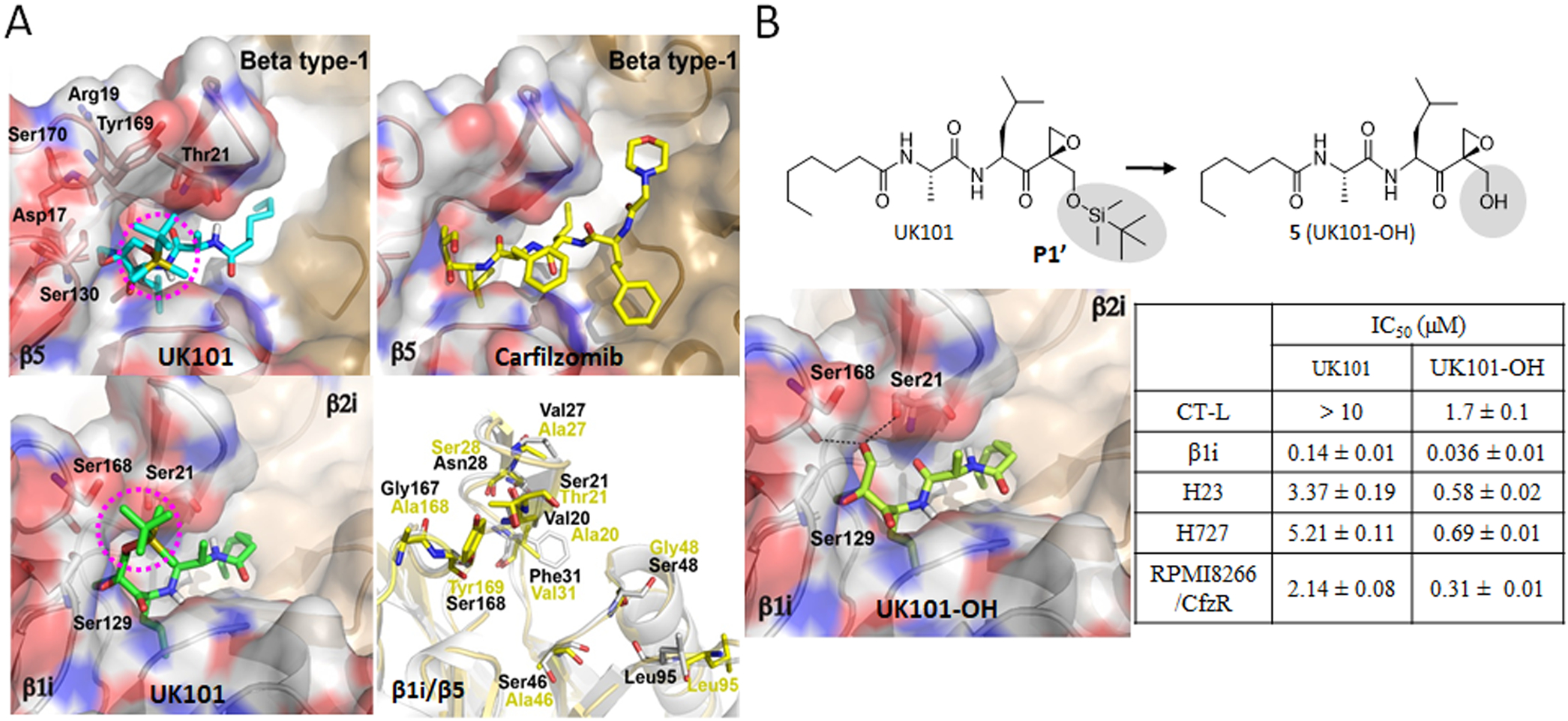
(A) Predicted docking models of UK101 and carfilzomib (Cfz) bound to β5 or β1i. The location of UK101’s TBDMS group positioned within putative P1′ pockets is highlighted using a purple-colored circle. β5 (PDB ID: 3UNB) and β1i (PDB ID: 3UNF) from mammalian 20S proteasomes were used as templates. In cartoon presentation, β1i (gray, PDB ID: 3UNF) was superposed to β5 (yellow, PDB ID: 3UNB) and only different amino acid residues are shown in stick model. (B) Comparison of UK101 and 5(UK101-OH) in terms of their potency (IC50 values) against proteasome chymotrypsin-like activity (in RPMI8226 cell lysate), β1i/LMP2 catalytic activity (in 20S purified human immunoproteasome), and against H23, H727, and Cfz-resistant RPMI8226 cells as measured by MTS cell viability assay. Data are reported as the mean ± SD. Docking model of UK101-OH bound to β1i (PDB ID: 3UNF). The P1′-OH of UK101-OH (5) is perfectly positioned to form hydrogen bonds with Ser168 and Ser21.
