Figure 3.
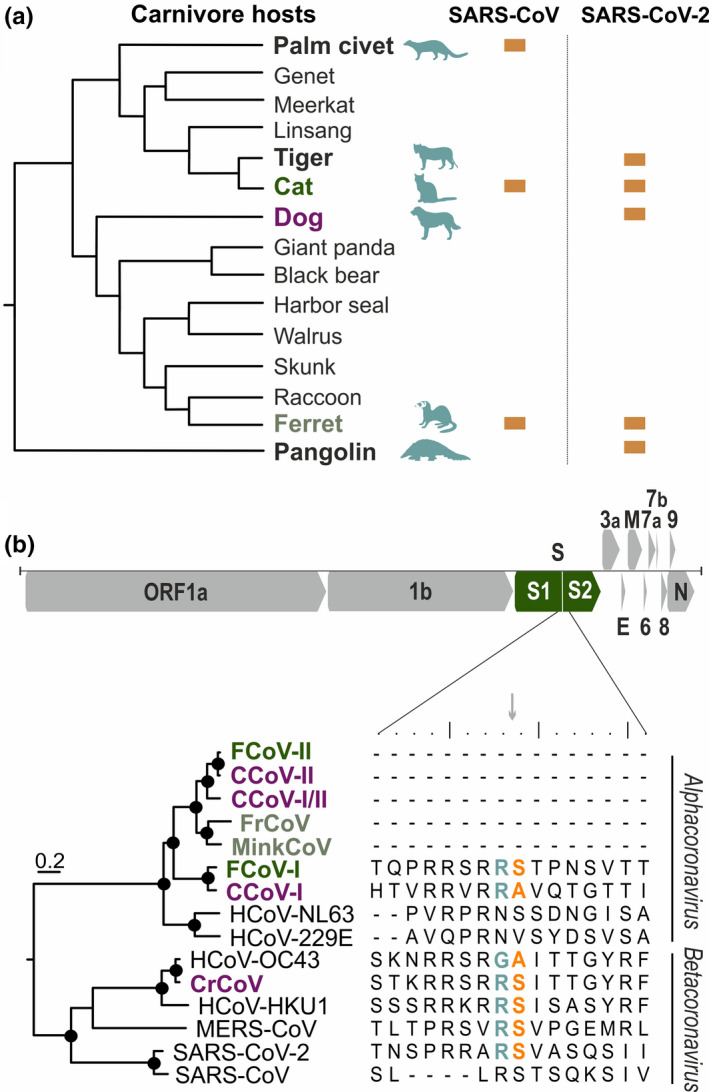
Susceptibility of carnivore hosts to coronaviruses (a) Carnivores susceptible to SARS‐related CoVs are dispersed across the family tree. Cladogram of carnivore families adapted from (Foley et al., 2016). (b) Furin cleavage site between the spike subunits S1 and S2 is predominantly present in betacoronaviruses. Scheme of SARS‐CoV‐2 genome organization with a magnified S1/S2 furin cleavage site within carnivore and human coronaviruses . Functional cleavage sites are highlighted with coloured ‘R S’ (orange and teal). Maximum‐likelihood tree of human and carnivore coronaviruses showing grouping of cat (green), dog (purple) and mustelid (grey) CoVs with human CoVs based on translated spike amino acid sequences. WAG + G + I was used as a substitution model and a complete deletion option was chosen. Scale bar indicates amino acid substitutions per site. Circle at nodes indicate bootstrap values ≥75% (1,000 bootstrap replicates). GenBank accession numbers: YP_004070194, AFG19726, AKZ66476, YP009256197, ADI80513, ACT10854, AEQ61968, YP_003767, NP_073551, AAR01015, AQT26498, YP_173238, YP_009047204, QHU36864, ACB69905 [Colour figure can be viewed at wileyonlinelibrary.com]
