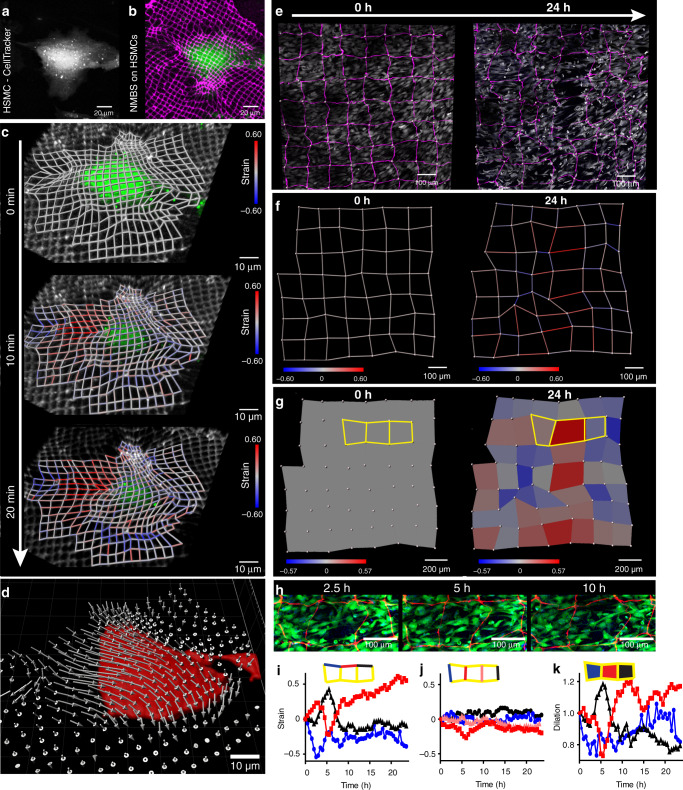
Fig. 3. 3D cellular strain mapping with the NMBS.
a Human skeletal muscle cells (HSMC) visualized with CellTracker-488. b Alexa-647 fibronectin NMBS (2 µm × 2 µm × 2 µm, magenta) applied to the HSMC (green). c Mapping of the NMBS strain over time reveals local regions of tension and compression following cell migration and contraction. d 3D displacement vector map (gray arrows) showing the magnitude and direction of displacement for all NMBS mesh nodes as the cell compresses (Red surface) over 20 min. e Alexa-555 fibronectin NMBS (100 µm × 100 µm × 10 µm, magenta) applied to C2C12 cells visualized with CellTracker-488 (gray). Images were acquired every 30 min for 24 h to track strain following transition to C2C12 differentiation media. f Strain mapping of the NMBS (red = tension, blue = compression) following transition to differentiation media reveals localized regions of tension and compression. g Area dilation was calculated for each section of the NMBS over time and shows spatial regions of expansion and contraction. h Regions of interest extracted showing cellular elongation events at 2.5, 5, and 10 h following transition to differentiation media (NMBS, red; C2C12 cells, green). i, j Quantification of segments from the region of interest in “g” during the C2C12 differentiation process shows region-specific strain rates during the 24-h time course, with the largest magnitude strains occurring in the x-direction. k Time-dependent mapping of regional dilation during the C2C12 differentiation process for ROI analyzed in (i, j).