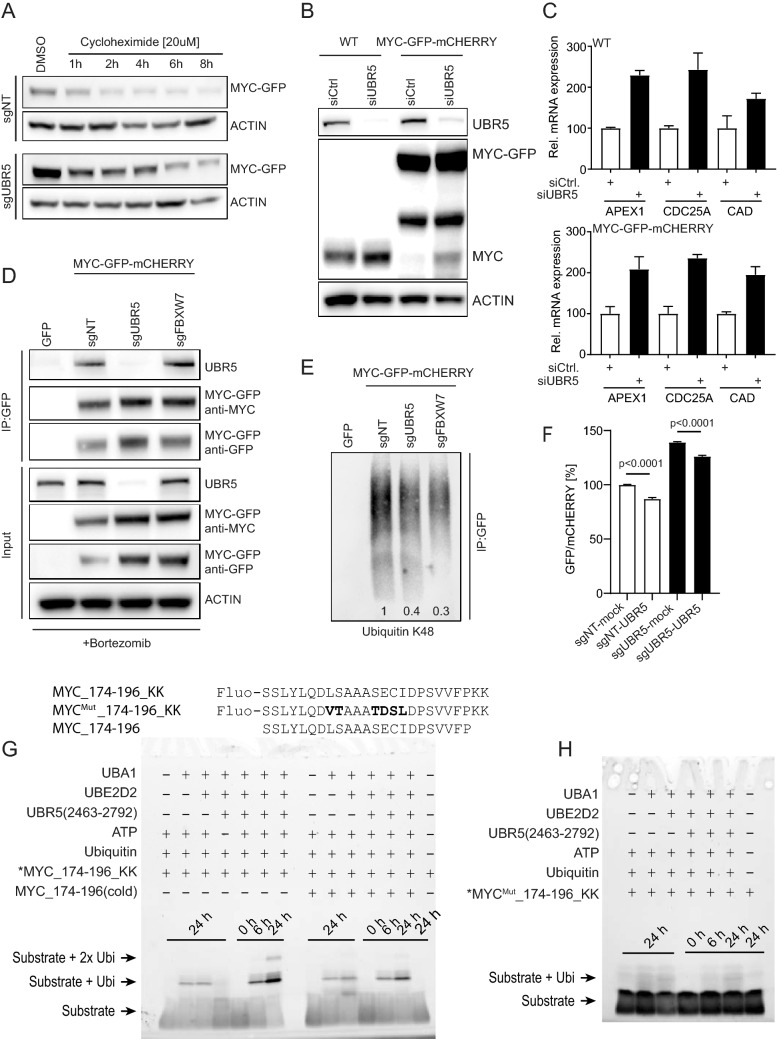
Figure 4.
Validation of UBR5 as a regulator of MYC protein stability and interaction. (A) B6 clone was transfected with sgNT or sgUBR5 for 7 days, followed by treatment with cycloheximide at 20 µM concentrations. MYC-GFP level were assessed by western blot at the indicated time points (quantification is shown in Supplementary Fig. 4A). (B) Increase in the protein level of MYC-wt as well as MYC-GFP fusion protein upon UBR5 knockdown in HEK293A cells (WT) and B6 clone (MYC-GFP-mCHERRY). (C) qPCR analysis of MYC target genes upon knockdown of UBR5. (D) Immunoprecipitation analysis showing interaction between UBR5 and MYC in B6 clone. (E) Decrease in ubiquitination of MYC-GFP immunoprecipitated protein upon knockout of UBR5 and FBXW7. Relative decrease in K48 ubiquitination was normalized to sgNT (1) and values are indicated on the bottom of the corresponding lane. (F) B6 clone was transfected with sgNT or sgUBR5 for 4 days and then further transfected with vector expressing mock DNA or UBR5 ORF, respectively to rescue the phenotype. GFP/mCHERRY was measured 72 h after transfection with rescue construct. (G) Ubiquitination assay utilizing UBR5(2463–2792) and peptide substrates MYC_174–196_KK (Fluo-SSLYLQDLSAAASECIDPSVVFPKK) and MYC_174–196 (SSLYLQDLSAAASECIDPSVVFP) resolved on a 5–20% SDS-PAGE gel. Fluorescein fluorophore was detected on a ChemiDoc System (Bio-Rad). (H) Ubiquitination assay utilizing UBR5(2463–2792) and the conservatively mutated peptide substrate MYC_174–196_KKMut (Fluo-SSLYLQDVTAAATDSLDPSVVFPKK), in which mutated residues are highlighted in bold. Samples were resolved on a 5–20% SDS-PAGE gel and fluorescence was detected on a ChemiDoc System (Bio-Rad).