Figure 5.
Functional and Evolutionary Constraint on Antibody-Escape Mutations
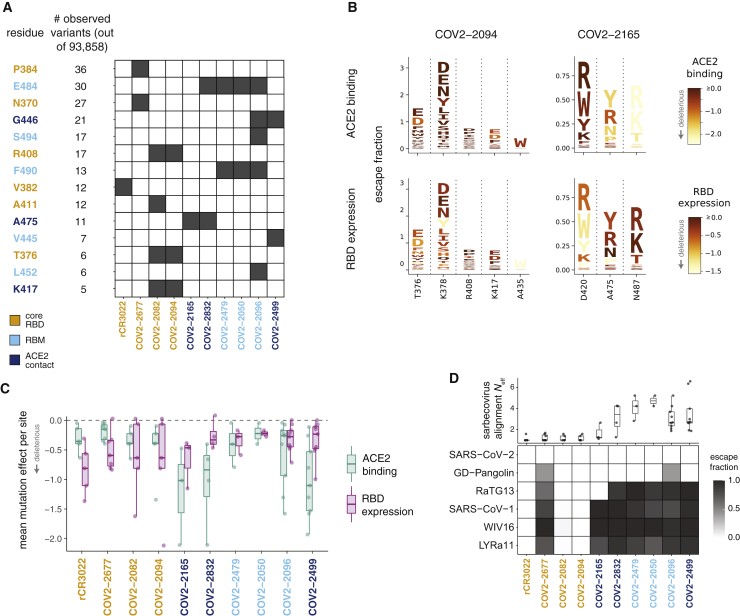
(A) Variation at sites of antibody escape among currently circulating SARS-CoV-2 viruses. For each site of escape from at least one antibody, we counted sequences in GISAID with an amino-acid change. Sites with at least 5 GISAID variants (of 93,858 sequences at the time of analysis) are shown ordered by count; black cells indicate antibodies with escape mutations at that site. Sites are colored by RBD region. Antibodies are colored according to where the majority of their sites of escape fall. See also Figure S4.
(B) Escape maps (as in Figure 2C), with letters colored according to how deleterious mutations are for ACE2 binding or RBD expression (Starr et al., 2020). Only sites of escape mutations for each antibody are depicted. See Figure S5 for similar logo plots for all antibodies.
(C) Mutational constraint on sites of escape. For each antibody, the mean effects of all possible amino acid mutations at sites of escape on ACE2 binding and RBD expression are shown.
(D) Top: effective number of amino acids (Neff) in the sarbecovirus RBD alignment at sites of escape for each antibody. Neff is a measure of the variability of a site (the exponentiated Shannon entropy), and ranges from 1 for a position that is completely conserved to 20 for a site where all amino acids are present at equal frequency. Bottom: escape fraction for each sarbecovirus RBD homolog from the yeast display selections; 1 means complete escape (no binding), and 0 means no escape (complete binding).