Figure 2.
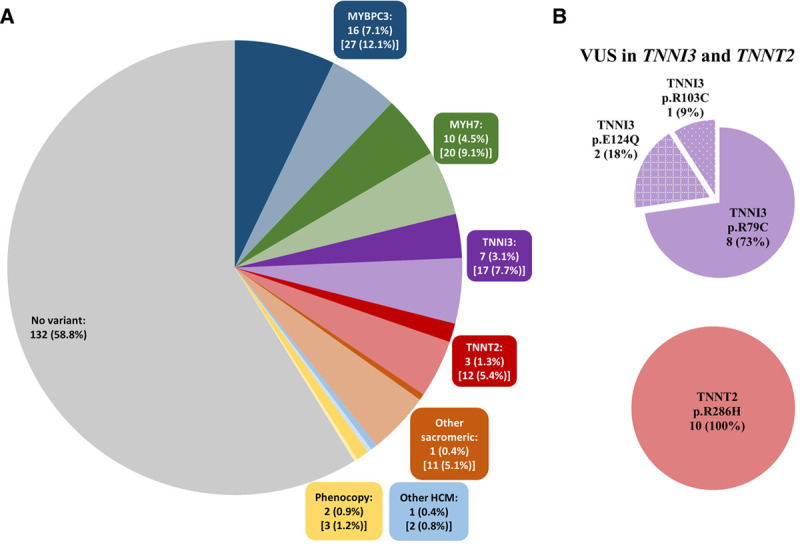
Pathogenic/likely pathogenic variants and excess variant of uncertain significance (VUS; exVUS = caseVUS%−controlVUS%) in hypertrophic cardiomyopathy (HCM) genes in Singaporean patients with HCM. A, Fifteen genes were assessed including major sarcomeric genes (MYBPC3, MYH7, TNNT2, and TNNI3), other sarcomeric genes (ACTC, MYL2, MYL3, TPM1, and TNNC1), other HCM genes (CSRP3, FHL1, and PLN), and geno/phenocopies (GLA, LAMP, and PRKAG2). The number and percentage refer to the total pathogenic/likely pathogenic (P/LP) case per gene (darker shade) while the number and percentage in parentheses refer to the total case excess of P, LP, and exVUS (lighter shade) as compared to genome aggregation database (gnomAD). B, The secondary pie charts show the proportion of all Singaporean HCM patients with TNNI3:p.R79C or TNNT2:p.R286H VUS as compared to other VUS in these genes, depicted overall in (A) by lighter shading.
