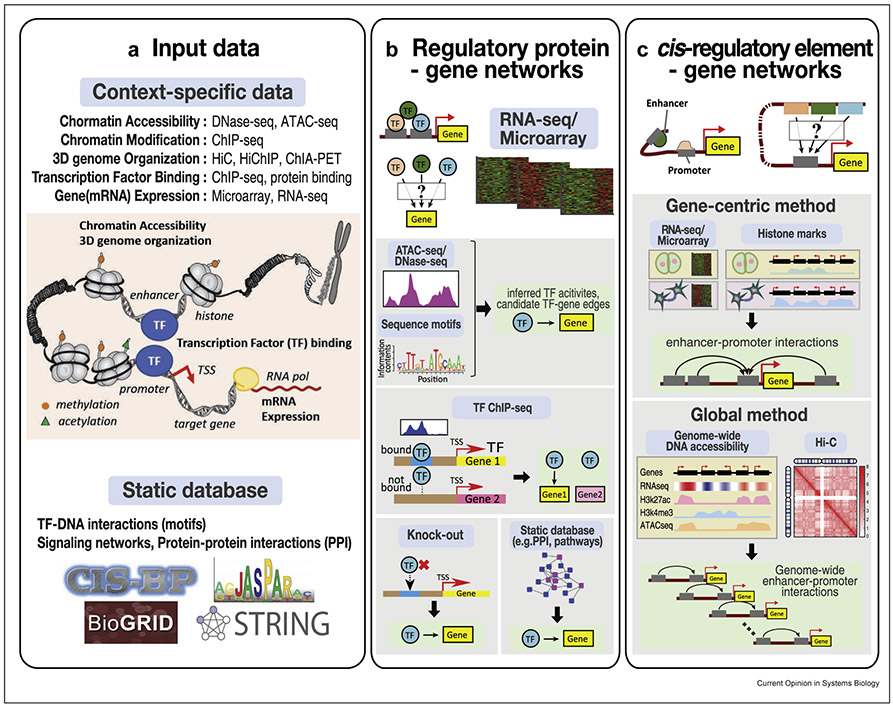
Figure 1. Data types used in integrative approaches for two main types of regulatory networks:
(a) Input data for inferring regulatory networks can be categorized into context-specific and static data sets. Context-specific gene regulatory events are measured across different layers of gene regulation using various genomic technologies. DNase-seq or ATAC-seq measure open regions of the chromatin and can be leveraged to identify regulatory elements. Chromosome conformation capture (3C) techniques such as Hi-C, HiChIP, and ChIA-PET allow global mapping of 3D genome organization. ChIP-seq and protein binding assays are used for detecting transcription factor (TF) binding to DNA. Transcriptomic assays measure expression levels of regulators and target genes under different conditions. Static databases often used in integrative inference of regulatory networks are DNA-binding motif collections such as JASPAR and CIS-BP, predefined signaling networks, and protein–protein interactions (PPIs). Databases of functional gene annotations and pathways are used for evaluating gene regulatory networks but can also be used as another source of data for integration. (b) Regulatory networks that capture the relationship between regulatory proteins, such as transcription factors and signaling proteins, and target genes. Gene expression measured with RNA-seq or microarrays is the primary data type for inferring these networks. Integrative inference approaches integrate other data types with gene expression such as sequence motifs, filtered by available accessibility (DNAse-seq, ATAC-seq) measurements, TF ChIP-seq, and gene perturbation data. (c) Cis-regulatory networks represent the noncoding regulatory sequences such as enhancers that regulate genes. These networks can be inferred either by learning a gene-centric model that predicts gene expression based on the chromatin state and/or accessibility of candidate enhancers across multiple conditions or a global model that learns predictive rules of enhancer–promoter interactions based on chromatin marks, architectural proteins, RNA-seq, and chromosome conformation (3C) data.