Fig 1. Schematic illustrating three ways that standard samples from COVID-19 clinical trials can be repurposed to assess the risk that vaccine resistance will evolve.
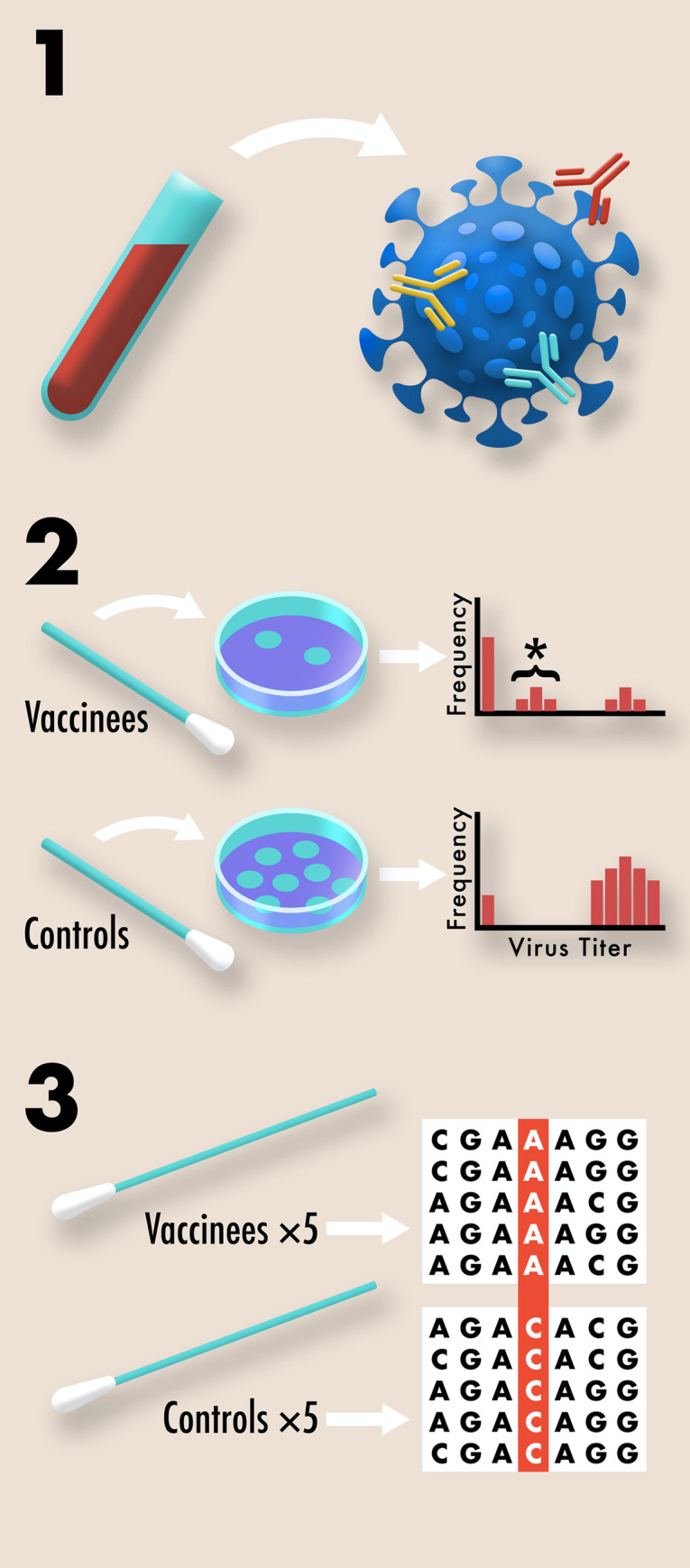
1. The complexity of B-cell and T-cell responses can be measured using blood samples [10,11]. Different neutralizing antibodies are depicted above in different colors. More complex responses indicate more evolutionarily robust immunity. 2. The effect of vaccination on transmission potential can be assessed by collecting viral titer data using routine nasal swabs. Plaque assays from multiple vaccinated and control individuals are compiled into a histogram. Undetectable viral titers suggest little or no transmission potential, due to either complete immune protection or the absence of exposure. High viral titers suggest high transmission potential due to the absence of a protective immune response. Intermediate viral titers, marked above with an asterisk, suggest moderate transmission potential due to partial vaccine protection. Intermediate titers indicate an increased risk for resistance evolution since pathogen diversity can be generated within hosts and selection can act during transmission between hosts. 3. Pre-existing variation for vaccine resistance can be assessed by recovering genome sequences from nasopharyngeal swabs of symptomatic COVID-19 cases included in the study. In a placebo controlled, double blind study, any significant differences in the genome sequences of samples from vaccinated and control individuals would suggest at least partial vaccine resistance.
