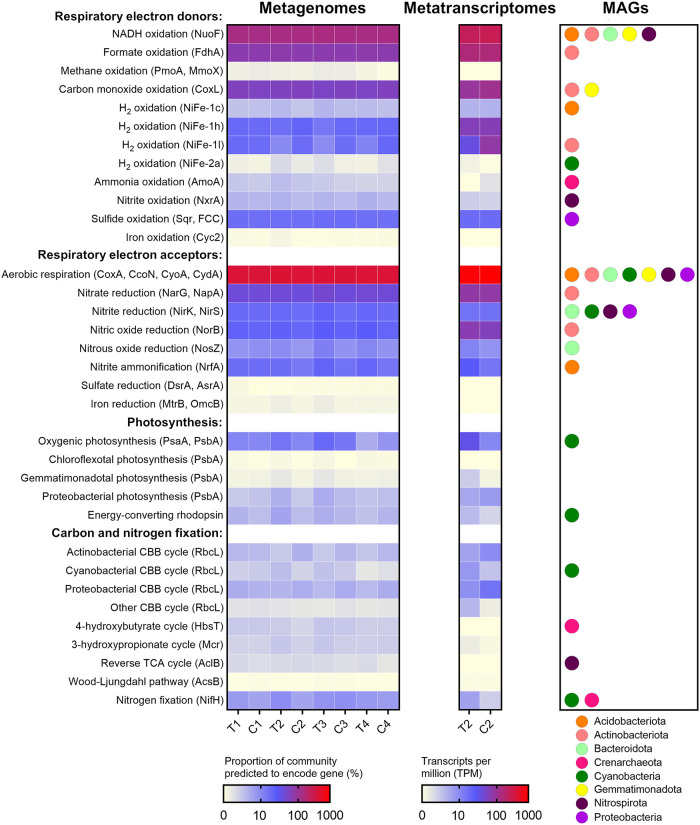
FIG 2.
Abundance, expression, and distribution of metabolic marker genes in the Australian desert soil microcosms. The left heat map shows the proportion of the total community predicted to encode each gene, based on normalizing metagenomic reads for these genes to single-copy ribosomal marker genes, across the four sampling points. Where multiple marker genes are present per category, the abundances are summed. Where community proportion exceeds 100%, each community member is predicted to carry more than one gene per category on average (e.g., multiple types of cytochrome oxidase). The middle heat map shows the abundance of each gene in the mapped metatranscriptomic reads, based on transcripts per million (TPM), for the first postwetting sampling point. The right plot shows the bacterial and archaeal phyla known to have each gene in the microcosms, based on the 39 metagenome-assembled genomes (MAGs). Note that given the moderate number and completeness of the MAGs, the phylum-level distribution of the genes is not exhaustive, and as a result, some abundant genes in the metagenomic short reads are not represented by any MAGs (e.g., group 1h [NiFe]-hydrogenases, type ID and IE RuBisCO, and proteobacterial photosystem II). Marker genes encoding subunits of the following enzymes involved in respiration, photosynthesis, and carbon and nitrogen fixation are shown: NuoF (complex I), FdhA (formate dehydrogenase), PmoA (particulate methane monooxygenase), MmoX (soluble methane monooxygenase), CoxL (type I [MoCu]-carbon monoxide dehydrogenase), NiFe ([NiFe]-hydrogenases, groups 1c, 1h, 1l, and 2a), AmoA (ammonia monooxygenase), NxrA (nitrite oxidoreductase), Sqr (sulfide-quinone oxidoreductase), FCC (flavocytochrome c sulfide dehydrogenase), Cyc2 (iron-oxidizing cytochrome), CoxA (cytochrome aa3 oxidase), CcoN (cytochrome cbb3 oxidase), CyoA (cytochrome bo3 oxidase), CydA (cytochrome bd oxidase), NarG (dissimilatory nitrate reductase), NapA (periplasmic nitrate reductase), NirK (copper-containing nitrite reductase), NirS (cytochrome bd1 nitrite reductase), NorB (nitric oxide reductase), NosZ (nitrous oxide reductase), NrfA (ammonifying nitrite reductase), DsrA (dissimilatory sulfite reductase), AsrA (anaerobic sulfite reductase), MtrB (decaheme iron reductase), OmcB (iron-reducing cytochrome), PsbA (photosystem I), PsbA (photosystem II), energy-converting rhodopsins, RbcL (ribulose 1,5-bisphosphate carboxylase/oxygenase), HbsT (thaumarchaeotal 4-hydroxybutyryl coenzyme A [CoA] synthase), Mcr (malonyl-CoA reductase), AclB (ATP-citrate lyase), AcsB (acetyl-CoA synthase), and NifH (nitrogenase).