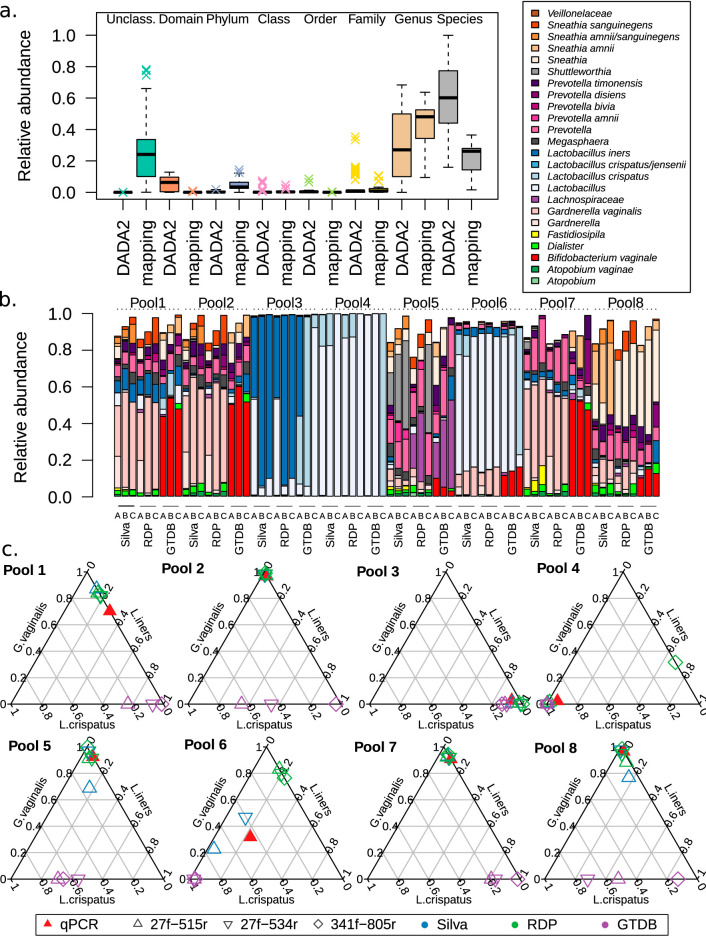
FIG 4.
Effects of various parameters on the taxonomic annotation of real amplicons. The taxonomic accuracy of each of the 16S primer sets is good, but V1-V3–534r yields more shallow annotations. It can, however, reliably detect Chlamydia trachomatis spike-in DNA. (a) Box plots showing the depth of classification for each sequence with different classification strategies. The DADA2 classifier yielded higher taxonomic resolution thatn simply mapping, regardless of the database used. (b) Taxonomy bar plots for each of the pools, processed with 2-step PCR with each of the primer sets and with each of the databases SILVA, RDP, and GTDB. An average for each triplicate is shown. Each technical replicate can be seen in Fig. S2. Only ASV with >10 counts are included in this figure. (c) Same samples as in panel a, compared to qPCR results for Lactobacillus iners, Lactobacillus crispatus, and Gardnerella vaginalis. For each sample, the sum of these three taxa was normalized to 1, to make them comparable to the qPCR results in the triaxial plot.