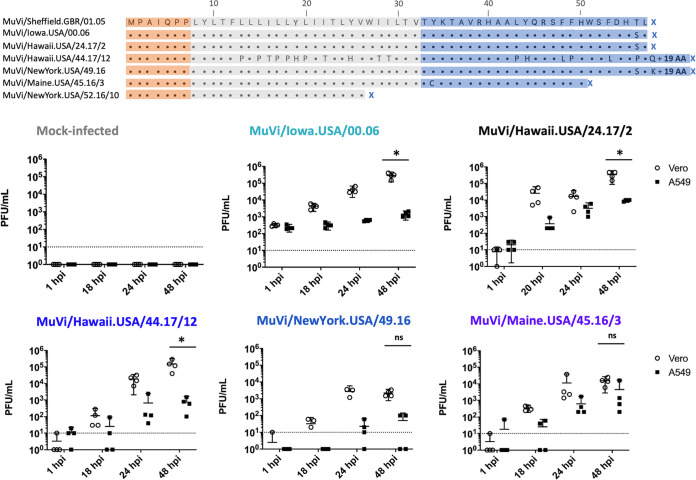
FIG 1.
(A) Amino acid alignment of a panel of contemporary, noncanonical SH amino acid sequences aligned against the SH amino acid sequence of the reference virus MuVi/Sheffield.GBR/01.05[G]. MuV SH protein topology is indicated by the superimposed shaded boxes; the ectodomain, transmembrane domain, and cytoplasmic domain are shown in orange, gray, and blue, respectively. (B) The replication kinetics of virus isolates bearing variant SH genes were evaluated through paired experiments at an MOI of 0.1 in Vero cells, which have interferon signaling defects and are routinely used for MuV isolation, and in interferon-competent A549 cells. Supernatant was sampled at the indicated time points and infectious virus titers from four biological replicates of each condition were determined for all samples by plaque assay on Vero cells. The limit of detection is indicated by a dashed line. Results shown are representative of three independent experiments. The well-characterized isolate MuVi/Iowa.USA/00.06 bearing a canonical SH sequence was included for comparison to contemporary viruses.*, P < 0.05, Vero versus A549; ns, not significant (two-way ANOVA, by virus, with post hoc Sidak correction for multiple comparisons using GraphPad Prism v8).