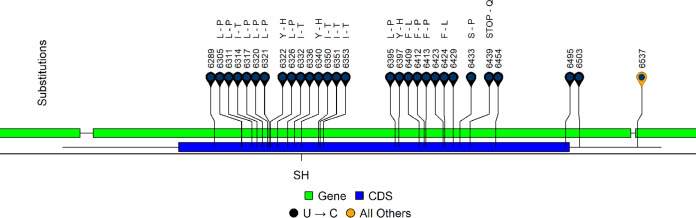
FIG 3.
SH substitutions observed in seven sequenced MuV strains bearing hypervariable SH genes, relative to MuVi/Iowa.USA/00.06. Strains were geographically and temporally similar to MuVi/Hawaii.USA/44.17/12, the source of noncanonical SH2 in Fig. 2B. All sequenced isolates shared the same substitutions in this region of the MuV genome. Substitutions were classified as synonymous or coding (amino acid substitution is noted); U-to-C substitutions are noted by color. For instances where multiple substitutions contribute to the coding result at a single codon, they were considered separately for all reported calculations.