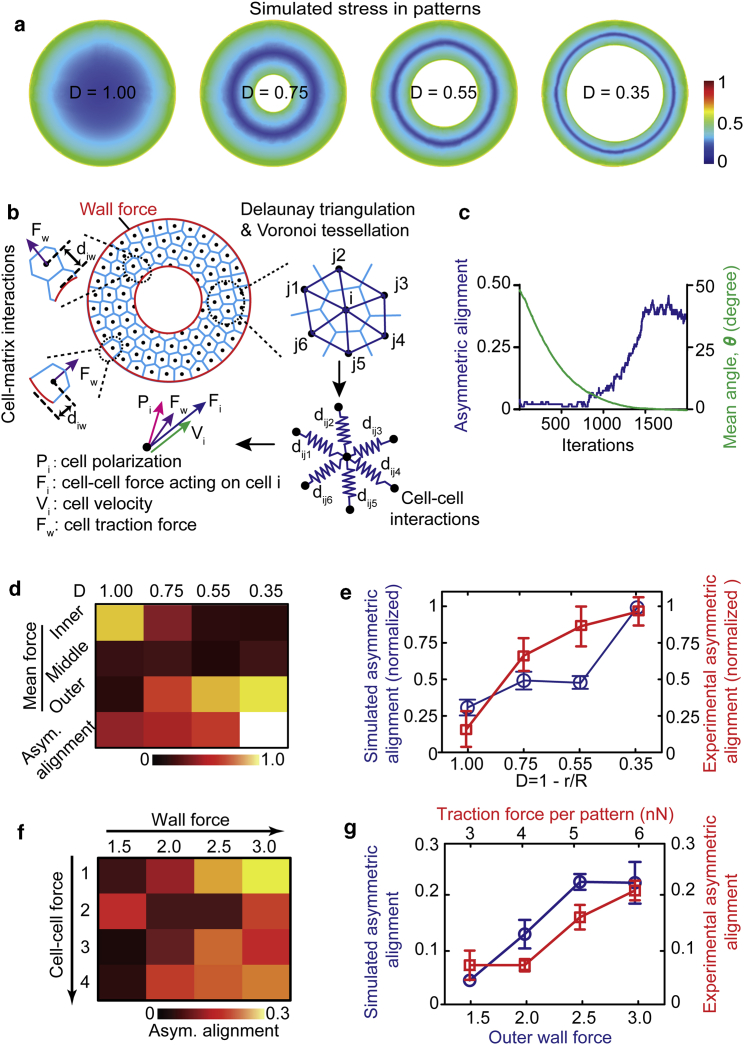
Figure 3.
Biophysical modeling of the force-dependent asymmetric cell alignment in the micropatterned vascular monolayer. (a) Simulated stress in different micropatterns is shown. The simulated micropatterns have the same sizes of the expertimental systems. The color coding represents the normalized magnitude of total von Mises stress on the bottom fixed surface, where 0 represents the minimal stress and 1 relates to the maximal stress. (b) A schematic shows an active particles model to simulate the micropatterned ring-shaped vascular sheets. Cells are represented as points (black dots) at their centers. Cell-cell interactions are mimicked by Delaunay triangulation (deep blue), which finds the closest nearby cell points and determines the forces (Fi) applied on the cell and the movement speed (Vi) accordingly. Voronoi tessellation (light blue) is used to visualize cells. Wall force is applied at the inner and outer boundaries of ring cell pattern (red circles) using exponential functions. Note that all parameters are nondimensional for all the simulations. (c) Quantified results are given, showing progression and stabilization of asymmetric alignments (blue line) as the mean angle (θ) of cells in the ring pattern (D = 0.55) decreasing to 0° (green line) along the iteration times. (d) Calculated total force magnitude and overall asymmetric alignments in different micropatterns (D = 1, 0.75, 0.55, 0.35) at iteration time of 2000 in the simulation model are given. It shows that the narrower the ring pattern with smaller value of “D,” the higher the asymmetric vascular cell alignment and relative force. Wall force was set as 0.5 for the inner wall and 3.0 for the outer wall. (e) Simulated (blue) asymmetric alignments in different patterns are consistent with the experimental (red) overall asymmetric alignments (normalized to maximum) in different micropatterns. (f) Calculated asymmetric alignment indexes from simulation models with different wall force (1.5–3.0) and cell-cell force (1–4) conditions in the ring patterns (D = 0.55) are shown. (g) Simulated results (blue) show the asymmetric cell alignment index correlates with the outer wall force in the ring pattern (D = 0.55, n ≥ 3), and experimental results (red) show the asymmetric cell alignment index correlates with the traction force magnitude in each pattern. In simulation, outer wall force was screened from 1.5 to 3.0 while keeping inner wall force constant as 0.5, and cell-cell force was kept consistent as 1. For the curve of the experimental results, each data point 3.0, 4, 5, 6 nN represents the group of traction forces per pattern in the range of 3–3.5, 1.5–4.5, 4.5–5.5, or 5.5–6.0 nN. Error bars represent ± standard errors. To see this figure in color, go online.