Extended Data Figure 4: Comparison of microbiota-host variable association strengths between disease-inclusive and disease-exclusive cohorts.
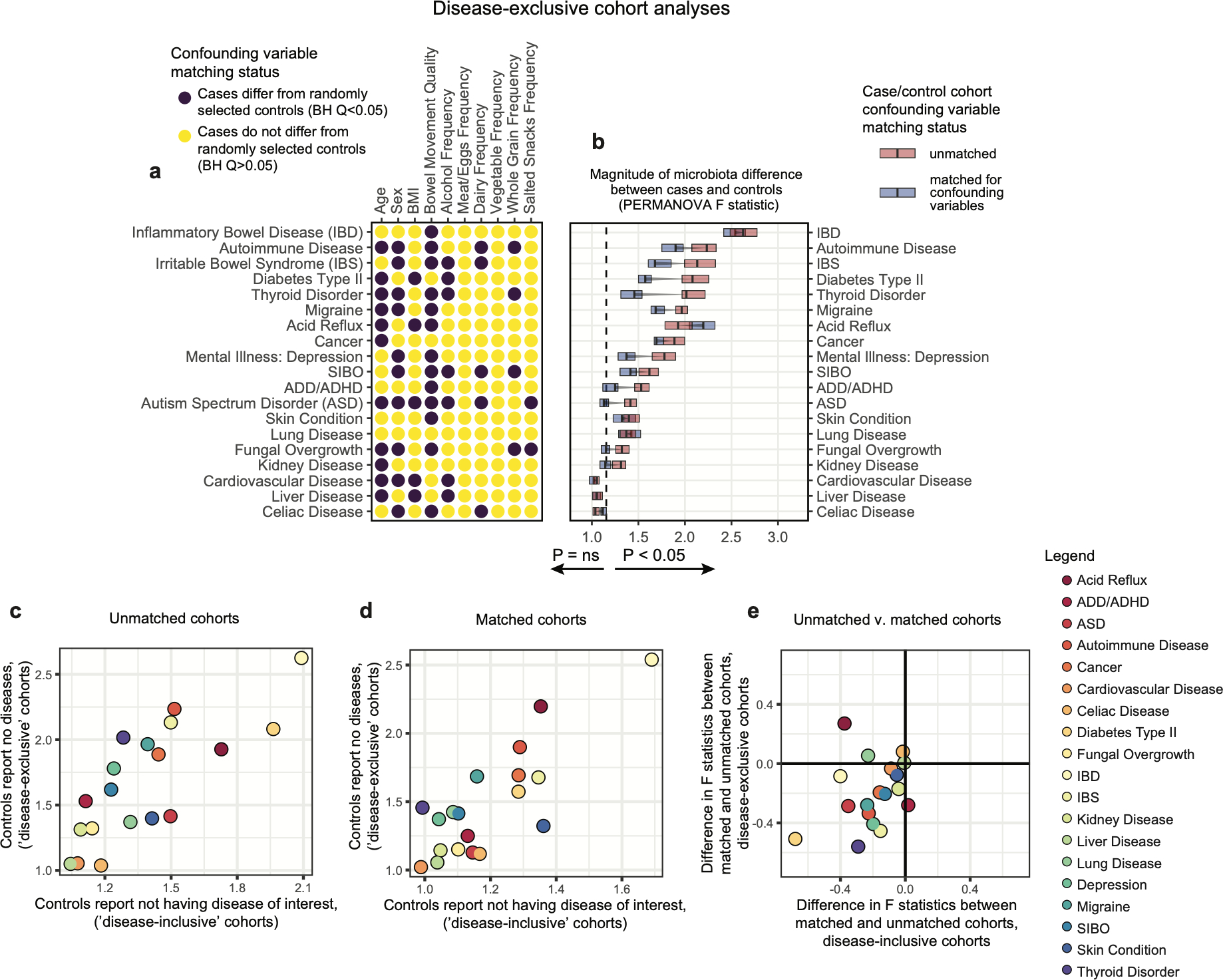
A-B) Differences in PERMANOVA F statistics between matched and unmatched cohorts within disease-exclusive analyses with all subjects reporting medical diagnoses removed, analogous to Figure 2. Subjects in ‘matched’ cohorts were matched for confounding variables shown to differ between cases and controls (purple) in panel A on a per-disease basis. Boxes represent interquartile ranges in F statistics from 25 permuted cohorts per matched/unmatched condition. Center lines within boxes represent median F statistic values. C) F statistics denoting differences between cases and controls for each disease among unmatched (location-only matched) cohorts comparing disease-exclusive to disease-inclusive results. Spearman rho= 0.81, P = 3.2*10−5. D) F statistics denoting differences between cases and controls for each disease among confounder-matched cohorts comparing disease-exclusive to disease-inclusive results. Spearman rho= 0.64, P = 2.9*10−3. E) Concordance in whether matching reduces or increases case-control microbiota differences were examined for disease-inclusive and disease-exclusive results. Differences in F statistics between matched and unmatched cohorts for each disease were calculated. Shown are F statistics differences for disease-inclusive cohorts (x-axis) and disease-exclusive cohorts (y-axis). Chi-square P = 0.0073, assuming random distribution of points across quadrants as the null hypothesis.
