Fig. 1 |. Cryo-EM reconstructions and atomic modelling of the VZV A- and C-capsids.
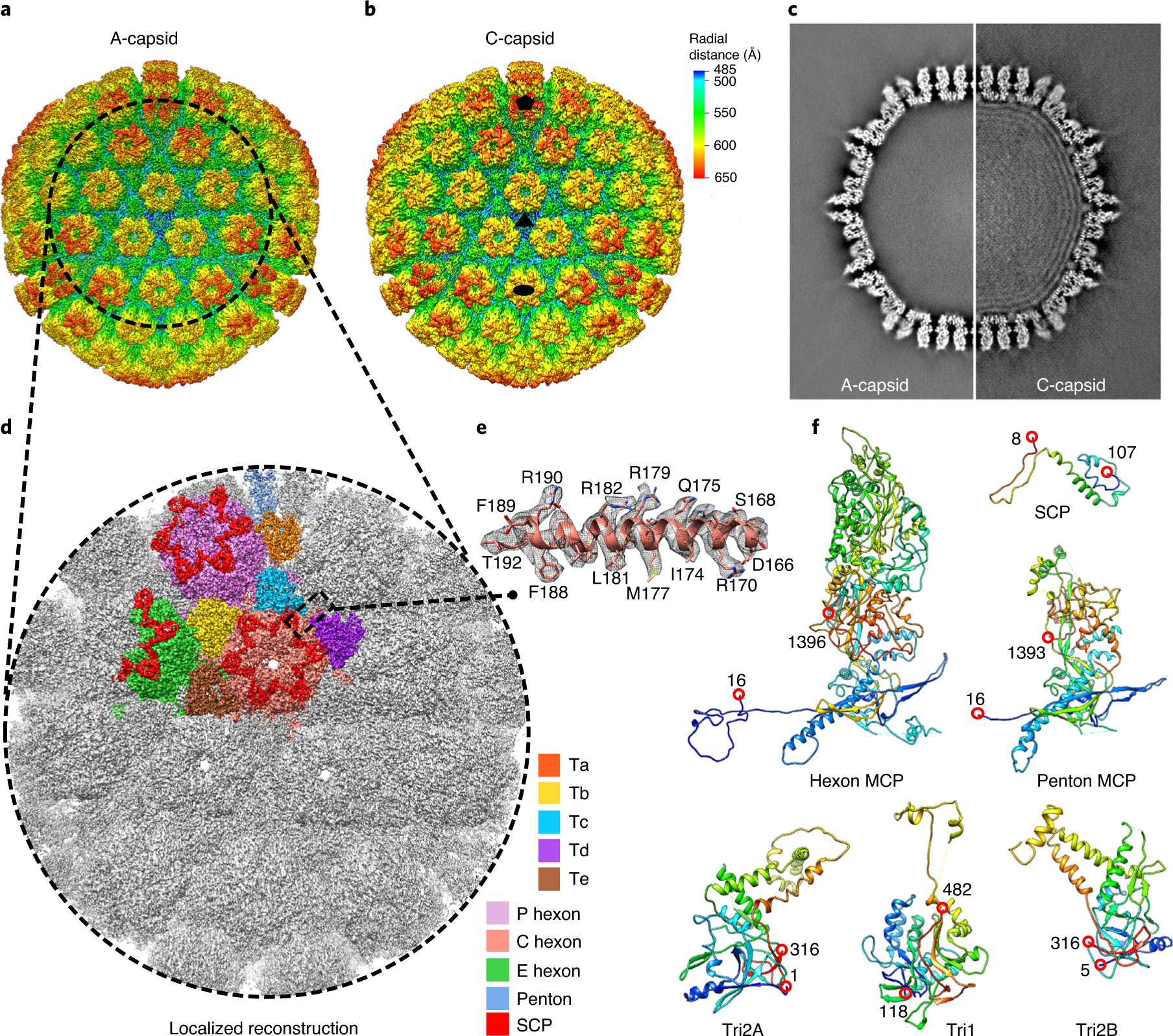
a,b, Radially coloured VZV A-capsid (a) and C-capsid (b) reconstructions at resolutions of 4.3 and 5.3 Å, respectively. Maps are viewed along a three-fold symmetry axis. The five-, three- and two-fold axes are denoted in the C-capsid reconstruction by a black pentagon, triangle and oval, respectively. c, Central slices of the VZV A-capsid (left) and C-capsid (right). The C-capsid slice shows interlayer dsDNA densities within the capsid shell. d,e, Localized reconstruction at 3.5 Å resolution of three-fold sub-particles in A-capsids (d) and a close-up view of the density map (grey mesh) of an α-helix in the MCP floor region, superposed with its model (e). f, Models of individual capsid proteins in ribbon representation in rainbow, from the N terminus (blue) to the C terminus (red).
