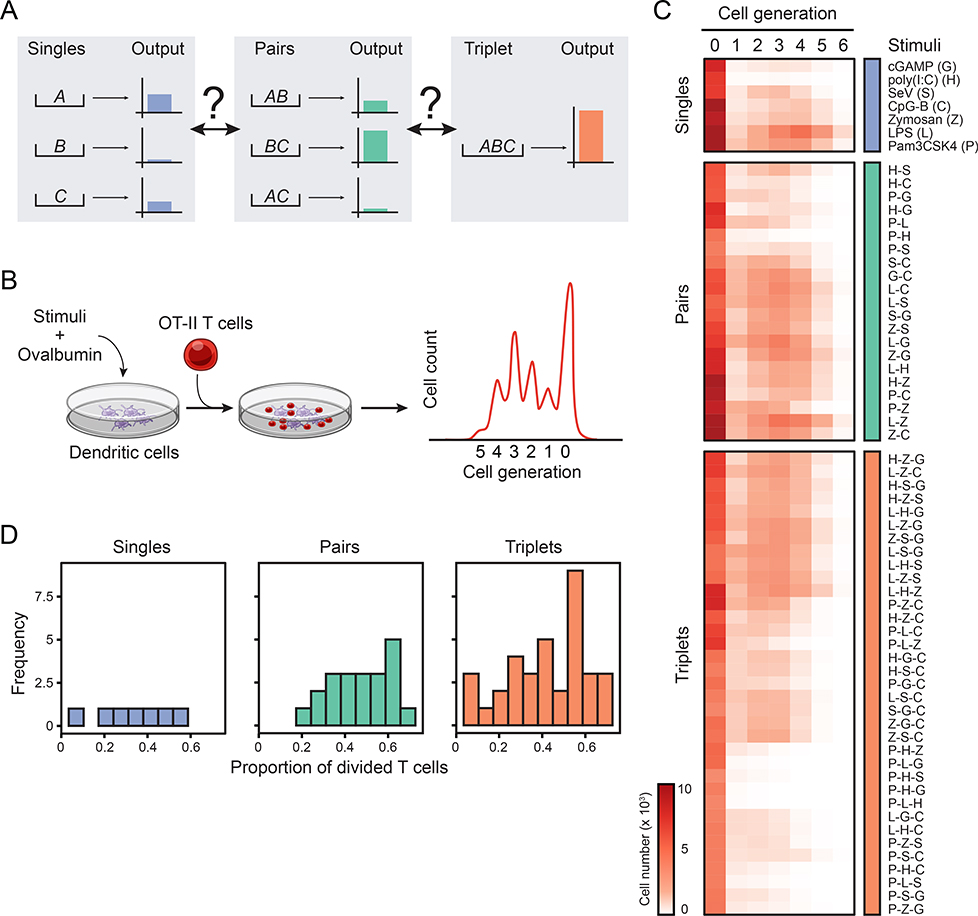
Figure 1. Systematic analysis of the combinatorial effects of microbial stimuli in a coculture system.
(A) Schematic depicting the unknown relationships between the effects of input singles (A, B andC; left), pairs (AB, AC and BC; center) and triplet (ABC; right) on a hypothetical output response.
(B) Schematic overview of the coculture assay used to measure the combinatorial effects of microbial inputs. Dendritic cells stimulated with singles, pairs or triplets of stimuli are pulsed with ovalbumin and subsequently incubated with transgenic OT-II T cells. T cell proliferation is measured by CFSE dilution.
(C) Heatmap showing all 63 microbial stimuli combinations (rows) used in the study: 7 singles (blue; top), 21 pairs (green; middle) and 35 triplets (orange; bottom). Columns indicate the cell generation and values are average cell numbers (color scale) (n = 8).
(D) Frequency distribution of the proportion of divided T cells for single (left), pair (center) and triplet (right) ligand stimulations.
See also Figures S1–2 and Table S1.