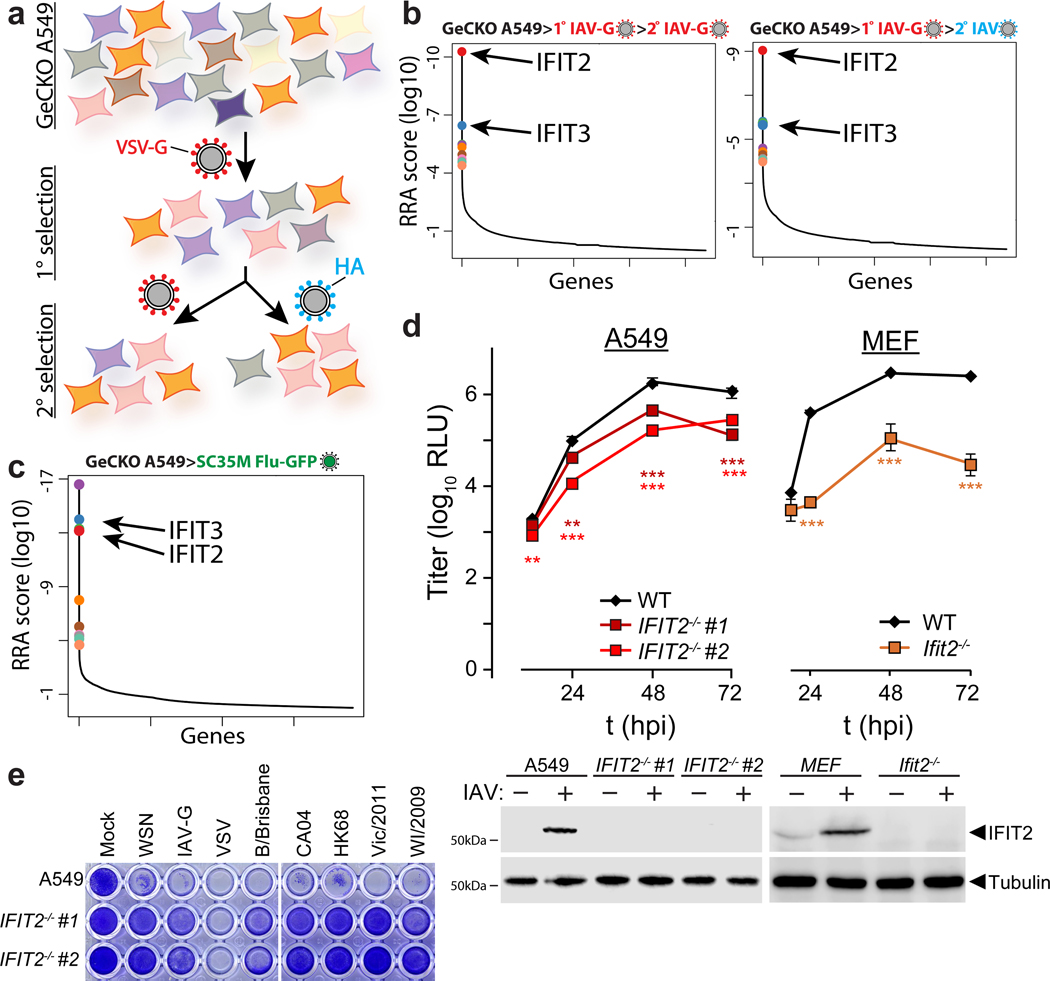
Figure 1. A CRISPR knockout screen identifies the antiviral proteins IFIT2 and IFIT3 as pro-viral regulators of influenza virus infection.
a, Schematic of the genome-scale CRISPR knockout (GeCKO) screen designed to specifically query host factors important for influenza virus infection after viral entry. b-c, MAGeCK (Model-based genome wide knock out)52 analysis for selection schemes involving sequential IAV-G selections (b, left), IAV-G followed by WSN (b, right), or seven rounds with SC35M Flu-GFP (c). d, Multicycle replication kinetics of WSN in wildtype or knockout cells. Western blotting confirmed loss of IFIT2 in two independent clonal human A549 cell lines and loss of Ifit2 in mouse embryo fibroblasts. Data are mean of n = 3 independent infections ± sd. (** p < 0.01, *** p < 0.001 when compared to WT; one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons or a two-tailed Student’s t-test). e, Viability of parental or IFIT2 knockout A549 cells following challenge with diverse primary influenza isolates and VSV. VSV-G; vesicular stomatitis virus glycoprotein. IAV; influenza A virus. IAV-G; IAV stably encoding VSV-G. SC35M; A/seal/Mass/1-SC35M/1980. WSN; A/WSN/1933. B/Brisbane; B/Brisbane/60/2008. CA04; A/California/04/2009. HK68; A/Hong Kong/1/1968. Vic/2011; A/Victoria/361/2011. WI/2009; A/Wisconsin/15/2009.