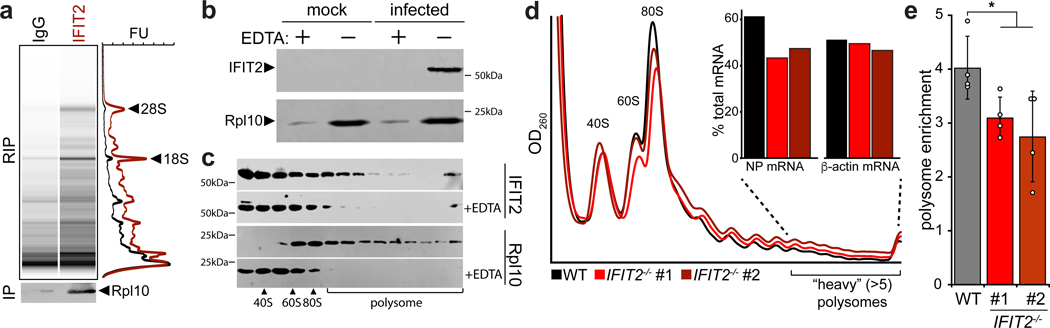
Figure 4. IFIT2 associates with active ribosomes and increases the translational efficiency of viral mRNAs.
a, Capillary electrophoresis traces for RNAs that co-precipitate with IFIT2 or an IgG control from WSN-infected cell lysate. Ribosomal RNAs were identified by their size and are indicated. Simulated gels and paired chromatographs quantified by fluorescence units (FU) are shown. In a separate IFIT2 immuno-precipitation, co-precipitating ribosomal protein (Rpl10) was detected by western blot. b-c, Translationally active ribosomes were separated from infected or mock cell lysate by polysome pelleting (b) or profiling (c). Ribosome were dissociated with EDTA where indicated. Proteins in polysome pellets or fractions were detected by western blot. d, Polysome profiles from WT and IFIT2−/− A549 cells infected with IAV WSN. mRNA abundance amongst highly translated messages (>5 ribosomes/mRNA) was determined by qRT-PCR (inset). e, Enrichment of NP mRNA in the polysome relative to total RNA was determined by qRT-PCR for samples from WT and IFIT2/- cells infected with IAV. Data are mean of n = 4 biologically independent samples +/− sd. (* p < 0.05, one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons to control conditions).