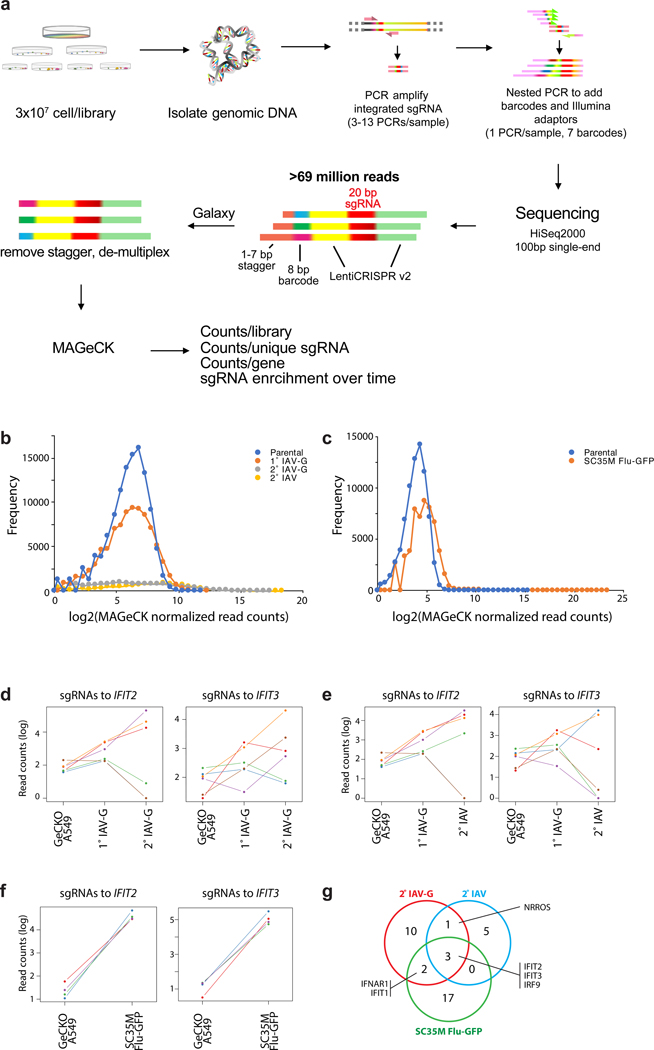
Extended Data Fig. 1. Enrichment of IFIT2 and IFIT3 over sequential influenza virus selections.
a, Schematic for hit analysis and selection. b-c, Frequency distribution of sgRNAs present in the parental libraries and those following selection with WSN (b) or SC35M (c). d-f, sgRNA enrichment trajectories following (d) sequential IAV-G selections, (e) IAV-G followed by bona fide influenza virus (WSN) infection or (f) selection with SC35M-GFP. The abundance of each sgRNA targeting the indicated gene is shown. g, Overlap between top hits from each screen. A conservative cutoff of RRA<10−5 was used to define top hits.