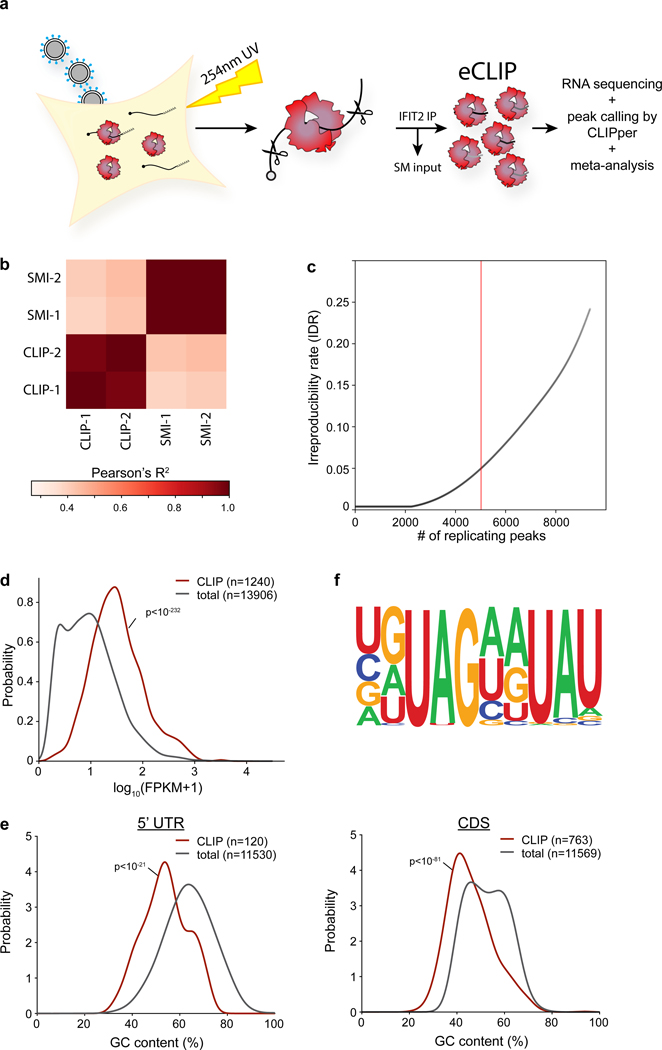
Extended Data Fig. 4. Analysis of IFIT2 CLIP-Seq data.
a, Experimental workflow for eCLIP of IFIT2 from IAV WSN-infected A549 cells. RNA-protein adducts were formed in infected cells by UV cross-linking. Lysates were prepared and RNA was partially digested with RNase. A fraction of lysate was removed to serve as the size-matched input control (SM input). IFIT2 was immunopurified and bound RNAs were processed via eCLIP followed by sequencing and analysis. b, CLIP-Seq was performed in duplicate and compared for IFIT2 CLIP or size-matched input (SMI) controls. Pearson’s correlation coefficient is shown as a heatmap. c, Concordance between ranked peaks in two independent biological replicates of IFIT2 CLIP-Seq was assessed by calculating an irreproducible discover rate (IDR). A conservative threshold of 5% was used to identify significant peaks. d, Abundance distribution in the size-matched input control for all transcripts and those with IFIT2 CLIP peaks. Populations were compared by a two-sided Mann-Whitney U-test. e, Meta-analysis of GC content for peaks identified in the 5’ UTR and coding sequence (CDS) of bound transcripts. IFIT2-bound RNAs were compared to all expressed 5’ UTRs or CDS via a two-sided Mann-Whitney U-test. f, A degenerate UAGnnUAU motif was found in ~20% of IFIT2 CLIP peaks (p = 10−279 compared to background).