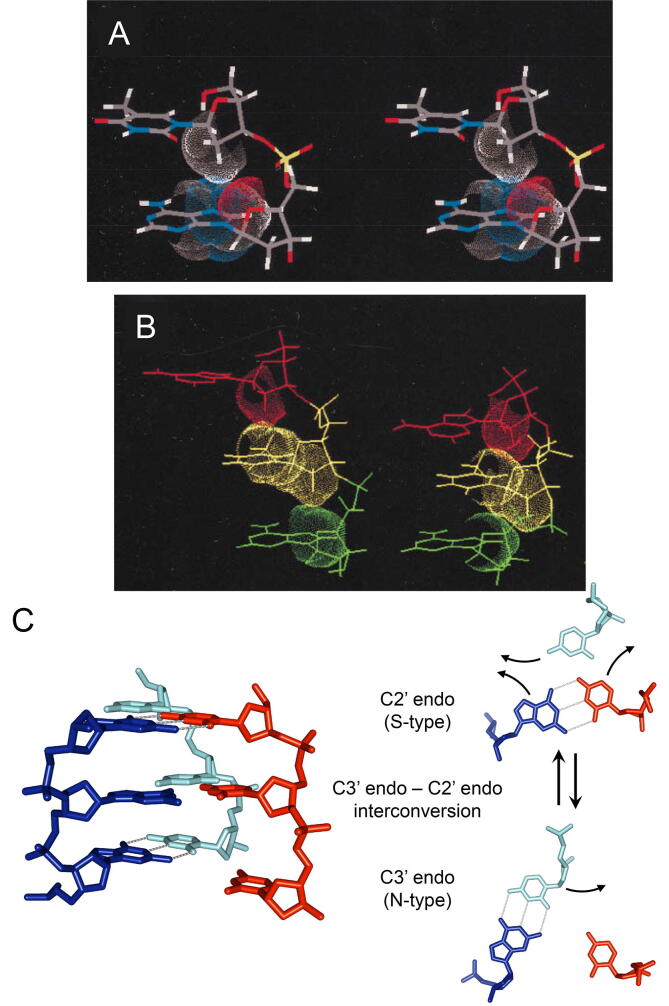
Fig. 4.
Extended DNA structure induced by binding to RecA/Rad51 (ATP-dependent) and ATP-independent homologous-pairing proteins, and a model for homology recognition by RecA/Rad51. A. Stereo view of extended ssDNA structure stabilized by CH-π interaction, an attractive molecular force [84], upon binding to RecA and Rad51 in the presence of an ATP-analogue [85], [86] and to ATP-independent homologous-pairing proteins [87]. Van der Waals spheres of a 2′-methylene moiety in a 5′ side residue (thymine) and of the base in the 3′ side residue (adenine) are indicated, and the contact of these surfaces represent CH-π interaction. B. Side views of extended DNA by binding to RecA (left) and B-form DNA (right) for a comparison. Van der Waals spheres of adjacent residues are indicated. In the extended DNA (left), in stead of base-base stacking, CH-π interaction stabilizes the distance between the planes of the adjacent bases (or axial rise per base) at nearly 5 Å, approximately 1.5 fold larger than that (3.5 Å) of B-form DNA (right), in which the structure is stabilized by base-base stacking. C. Model for homology search and W-C base-pairing between dsDNA extended by CH-π interaction and ssDNA (extended by binding to RecA). The 2D model (on the right) is the view from the bottom of the 3D model (on the left). Bases of dsDNA (dark and light blue, bottom) are randomly flipped out by the interconversion of sugar puckers from the C3′ endo to the C2′ endo (middle). An ssDNA (red) approaches the minor groove of the dsDNA (bottom). Only if the flipped base (dark blue) is complementary to the base of the ssDNA, the base-pair switch occurs (top) and the replace base (light blue) is in the major groove of the hybrid base pair that is newly formed. When this base-pair switch takes place between complementary sequences of dsDNA and ssDNA, homologous recognition and homologous-complex-core formation are accomplished simultaneously, and the replaced strand is left in the major groove of the newly formed hybrid duplex (See Fig. 2A). Panels A and B were reproduced with minor modification from Ref. [85], with permission (Copyright 1997, National Academy of Sciences, U.S.A.). Panel C is the reproduction of the Fig. 4 in Ref. [86], with permission (Copyright 1998, National Academy of Sciences, U.S.A.), generated using the coordinates of PDB code 1I1V (Model Archive DOI: https://doi.org//10.2210/pdb1I1V/pdb [86]). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)