Abstract
The nucleocapsid (N) protein of coronaviruses serves two major functions: compaction of the RNA genome in the virion and regulation of viral gene transcription. It is not clear how the N protein mediates such distinct functions. The N protein contains two RNA-binding domains surrounded by regions of intrinsic disorder. Phosphorylation of the central disordered region promotes the protein’s transcriptional function, but the underlying mechanism is not known. Here, we show that the N protein of SARS-CoV-2, together with viral RNA, forms biomolecular condensates. Unmodified N protein forms partially ordered gel-like condensates and discrete 15-nm particles based on multivalent RNA-protein and protein-protein interactions. Phosphorylation reduces these interactions, generating a more liquid-like droplet. We propose that distinct oligomeric states support the two functions of the N protein: unmodified protein forms a structured oligomer that is suited for nucleocapsid assembly, and phosphorylated protein forms a liquid-like compartment for viral genome processing.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, nucleocapsid, N protein, phosphorylation, biomolecular condensate, phase separation
Graphical Abstract
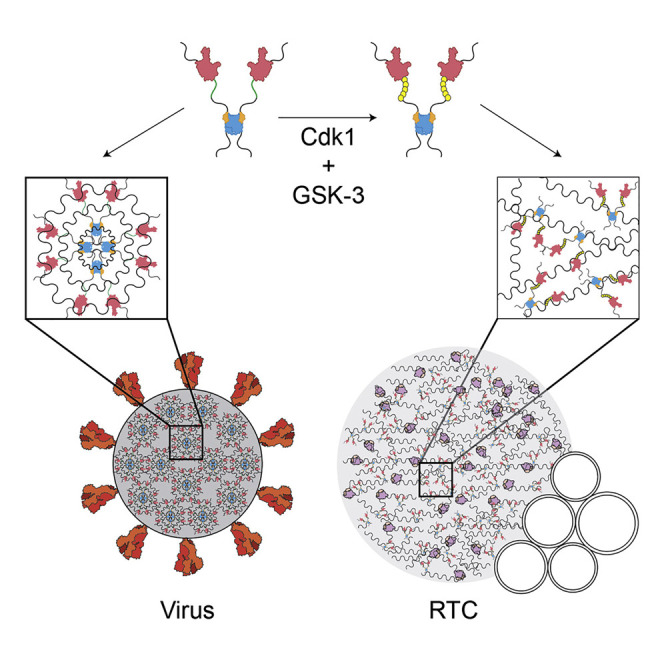
Carlson et al. demonstrate that the nucleocapsid (N) protein of SARS-CoV-2, together with viral RNA, forms gel-like biomolecular condensates and particles that are consistent with its genome-packaging role. Phosphorylation transforms condensates into liquid-like droplets, which may provide a cytoplasmic compartment to support the protein’s function in viral genome transcription.
Introduction
Coronaviruses are enveloped viruses with an ∼30-kb positive-sense single-stranded RNA genome, packed tightly inside the ∼100-nm virion in a poorly defined structure called the nucleocapsid. Condensation of the genome into the compact nucleocapsid structure depends primarily on the nucleocapsid (N) protein, one of the four major structural proteins encoded in the viral genome (Fung and Liu, 2019; Laude and Masters, 1995; Masters, 2006).
Following viral entry and disassembly of the nucleocapsid, the genome is translated to produce RNA-dependent RNA polymerase and numerous other non-structural proteins (Nsps) (Fung and Liu, 2019; Kim et al., 2020; Snijder et al., 2016; Sola et al., 2015). These proteins rearrange membranes of the endoplasmic reticulum to form the replication transcription complex (RTC) (den Boon and Ahlquist, 2010; Hagemeijer et al., 2012; Knoops et al., 2008; Neuman et al., 2014; Snijder et al., 2020; Stertz et al., 2007; Ulasli et al., 2010; Wolff et al., 2020), which is thought to provide a scaffold for the viral proteins that perform genome replication and transcription and which might also shield these processes from the host cell’s innate immune response.
Using genomic (+) RNA as a template, the viral polymerase produces (−) RNA transcripts of subgenomic regions encoding the viral structural proteins (S, E, M, and N) (Fung and Liu, 2019; Kim et al., 2020; Snijder et al., 2016; Sola et al., 2015). Subgenomic transcription involves a template-switching mechanism in which the polymerase completes transcription of a structural protein gene and then skips to a transcription-regulating sequence (TRS) at the 5′ end of the genome, resulting in subgenomic (−) RNA fragments—which are then transcribed to produce (+) RNA for translation. N protein is encoded by the most abundant subgenomic RNA and is translated at high levels early in infection. The N protein is the most abundant viral protein in the infected cell (Bouhaddou et al., 2020; Laude and Masters, 1995; Masters, 2006) and accumulates in dynamic clusters at RTCs (Bost et al., 2000; Cong et al., 2020; Snijder et al., 2020; Stertz et al., 2007; Ulasli et al., 2010; V’kovski et al., 2019; Verheije et al., 2010; Verheije et al., 2008) where it is thought to help promote the RNA structural rearrangements required for subgenomic transcription (Almazán et al., 2004; Wu et al., 2014; Zúñiga et al., 2010).
The structural features of the N protein are well conserved among coronaviruses. The ∼46-kDa N proteins of SARS-CoV and SARS-CoV-2 are ∼90% identical. The N protein contains two globular domains, the N- and C-terminal domains (NTD and CTD, respectively), surrounded by intrinsically disordered regions (Chang et al., 2014) (Figure 1 A, Figure S1). N protein is highly basic (pI ∼10), and multiple RNA-binding sites are found throughout the protein (Chang et al., 2009). The NTD is an RNA-binding domain (Fan et al., 2005; Huang et al., 2004; Jayaram et al., 2006; Kang et al., 2020). The CTD forms a tightly linked dimer with a large RNA-binding groove (Chen et al., 2007; Takeda et al., 2008; Ye et al., 2020; Zuwala et al., 2015), and the fundamental unit of N protein structure is a dimer (Luo et al., 2004; Yu et al., 2005). Under some conditions, the dimer self-associates to form oligomers that depend on multiple protein regions (Chang et al., 2013; Chen et al., 2007; Cong et al., 2017; He et al., 2004; Hurst et al., 2009; Luo et al., 2006; Luo et al., 2005; Ye et al., 2020; Zuwala et al., 2015). The biochemical features and function of these oligomers are not clear.
Figure 1.
SARS-CoV-2 N Protein Forms Biomolecular Condensates in the Presence of RNA
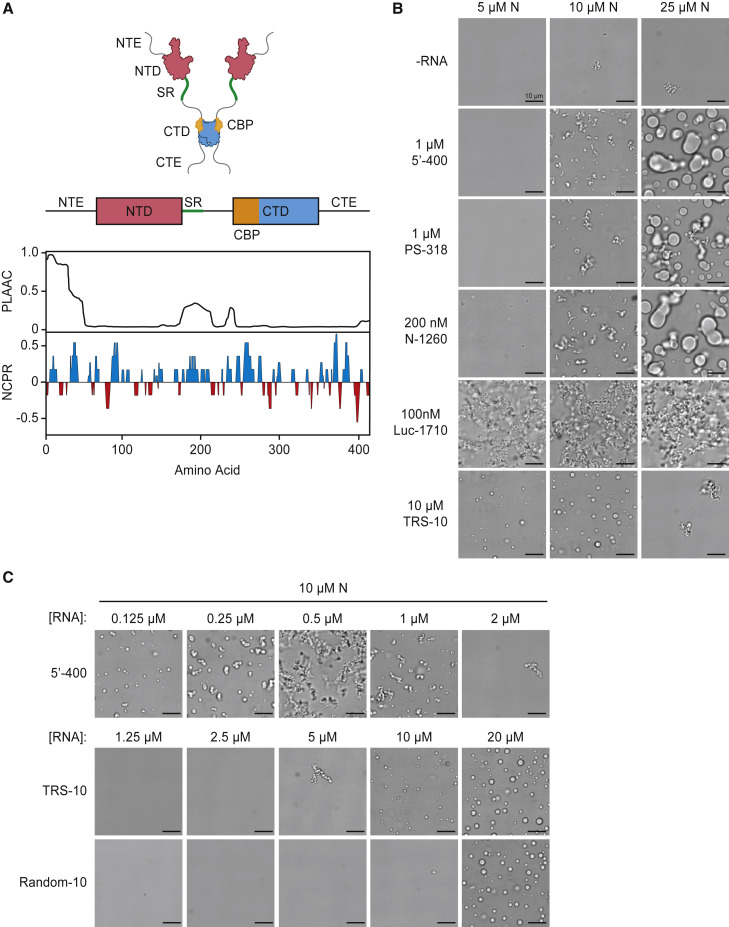
(A) Top: schematic of N protein domain architecture. NTE, N-terminal extension; NTD, N-terminal domain; SR, SR region; CTD, C-terminal domain; CTE, C-terminal extension; CBP, CTD basic patch. Bottom: features of amino acid sequence. PLAAC, prion-like amino acid composition (Lancaster et al., 2014); NCPR, net charge per residue. See Figure S1 for sequence.
(B) Light microscopy images of N protein condensates after a 30-min incubation at room temperature with the indicated RNA molecules. All images are representative of multiple independent experiments; scale bar, 10 μm.
(C) Condensate formation by N protein (10 μM) over a range of 5′-400, TRS-10, and Random-10 RNA concentrations. All images are representative of multiple independent experiments; scale bar, 10 μm. Random-10 RNA is a mixed population of 10-nt random sequences. See Figure S3 for condensate formation at other N protein and 5′-400 RNA concentrations.
The central disordered linker contains a conserved serine-arginine (SR)-rich sequence that is likely to serve as a key regulatory hub in N protein function. Early in infection, the SR region is rapidly phosphorylated at multiple sites by cytoplasmic kinases (Bouhaddou et al., 2020; Davidson et al., 2020; Fung and Liu, 2018; Klann et al., 2020; Peng et al., 2008; Stohlman et al., 1983; Surjit et al., 2005; White et al., 2007; Wu et al., 2014; Wu et al., 2009). Phosphorylation leads to association with the RNA helicase DDX1, which promotes RNA structural changes required for transcription of long subgenomic RNAs in the RTC (Wu et al., 2014). Later in infection, nucleocapsid formation and viral assembly do not seem to require N protein phosphorylation, which is substantially reduced in the nucleocapsid of MHV and SARS-CoV virions (Wu et al., 2014; Wu et al., 2009). We have little understanding of the molecular mechanisms by which phosphorylation influences N protein function.
To gain a better understanding of N protein function and its control by phosphorylation, we explored the biochemical properties of the N protein from SARS-CoV-2, the causative agent of the ongoing COVID-19 pandemic. We found that N protein and viral RNA form biomolecular condensates (Alberti et al., 2019; Banani et al., 2017; Owen and Shewmaker, 2019; Shin and Brangwynne, 2017; Tauber et al., 2020). The ordered, gel-like condensates of the unmodified protein are changed to more liquid-like droplets by phosphorylation, providing a potential structural foundation for the distinct functions of the N protein in nucleocapsid assembly and genome processing.
Results
N Protein of SARS-CoV-2 Forms RNA-Dependent Biomolecular Condensates
Purified N protein produced in bacteria migrated on gel filtration as a dimer in high salt but as a large oligomer in physiological salt (Figure S2A). Light microscopy revealed the presence of liquid-like droplets in the presence and absence of added RNA (Figure S2B), suggesting that N protein forms biomolecular condensates. We noted, however, that purified N protein contained nucleic acid, even after nuclease treatment, raising the possibility that structures seen in the absence of added RNA were due to tightly bound contaminating RNA. Following removal of RNA by protein denaturation and renaturation (Figure S2C), the protein displayed few microscopic structures, but addition of viral RNA greatly enhanced the formation of structures similar to those in the native preparation (Figure 1B). We conclude that RNA is required for the formation of the higher-order oligomers seen in the microscope. All subsequent studies were performed with denatured and renatured proteins (Figure S2D).
We analyzed the effects of three mid-sized viral RNA fragments: (1) 5′-400, containing the 400 nt at the 5′ end of the SARS-CoV-2 genome, which is thought to include multiple secondary structure elements and the leader TRS (Chen and Olsthoorn, 2010; Yang and Leibowitz, 2015; Yang et al., 2015); (2) PS-318, a 318-nt sequence near the end of ORF1b in SARS-CoV, proposed as a packaging sequence (Hsieh et al., 2005; Woo et al., 2019) but of unknown function (Chen and Olsthoorn, 2010; Masters, 2019); and (3) N-1260, a 1260-nt sequence containing the open reading frame of the N gene near the 3′ end of the SARS-CoV-2 genome. RNA encoding firefly luciferase (Luc-1710) was a nonviral control. N protein structures were analyzed by microscopy 30 min after addition of RNA at room temperature (Figure 1B). At 10 μM N protein, the three viral RNAs rapidly generated branched networks of liquid-like beads. Higher N protein concentrations produced large liquid-like droplets several microns in diameter. Nonviral RNA led to amorphous filamentous aggregates with partial liquid-like appearance. For most subsequent studies, we used 5′-400 RNA as a representative viral RNA.
Incubation at higher temperature (30°C or 37°C) had little effect on droplet formation in a 30-min incubation (Figure S2E), and longer incubations did not transform filamentous networks into droplets (Figure S2F). High salt dissolved N protein structures, indicating that they depend primarily on electrostatic interactions (Figure S2G).
We analyzed the formation and morphology of condensates over a wide range of N protein and 5′-400 RNA concentrations (Figures S3 and 1C, top row). At low RNA concentrations, small spherical droplets were seen at multiple N protein concentrations. Higher RNA and protein concentrations led to filamentous structures. Interestingly, at RNA concentrations approaching those of N protein, no structures were formed. These results suggest that condensates depend on the crosslinking of multiple N proteins by a single RNA.
We tested the importance of multivalent RNA binding by measuring the effects of a 10-nt RNA carrying the TRS sequence of SARS-CoV-2. The TRS sequence is thought to bind primarily to the NTD, with some contribution from the adjacent SR region (Grossoehme et al., 2009; Keane et al., 2012). Surprisingly, addition of the TRS RNA triggered the rapid formation of droplets without the filamentous structures seen with longer viral RNAs (Figures 1B and 1C). Also in contrast to results with longer RNAs, droplet formation was greatly reduced when the N protein was in molar excess over the TRS RNA (Figures 1B and 1C), indicating that RNA-free N protein exerts a dominant inhibitory effect on the formation of TRS-bound oligomers. Droplets formed when TRS RNA was equimolar with or in excess of N protein (Figure 1C), suggesting that these droplets depend on the binding of a single TRS RNA to each N protein. A 10-nt RNA with a random sequence displayed similar behavior but at higher concentrations, suggesting that specific binding is not entirely responsible for the effects of TRS RNA (Figure 1C). We speculate that the binding of a short RNA molecule alters N protein structure to promote low-affinity protein-protein interactions leading to droplet formation. In the more physiologically relevant context of long RNAs, these weak protein-protein interactions are presumably augmented by multivalent RNA-protein interactions.
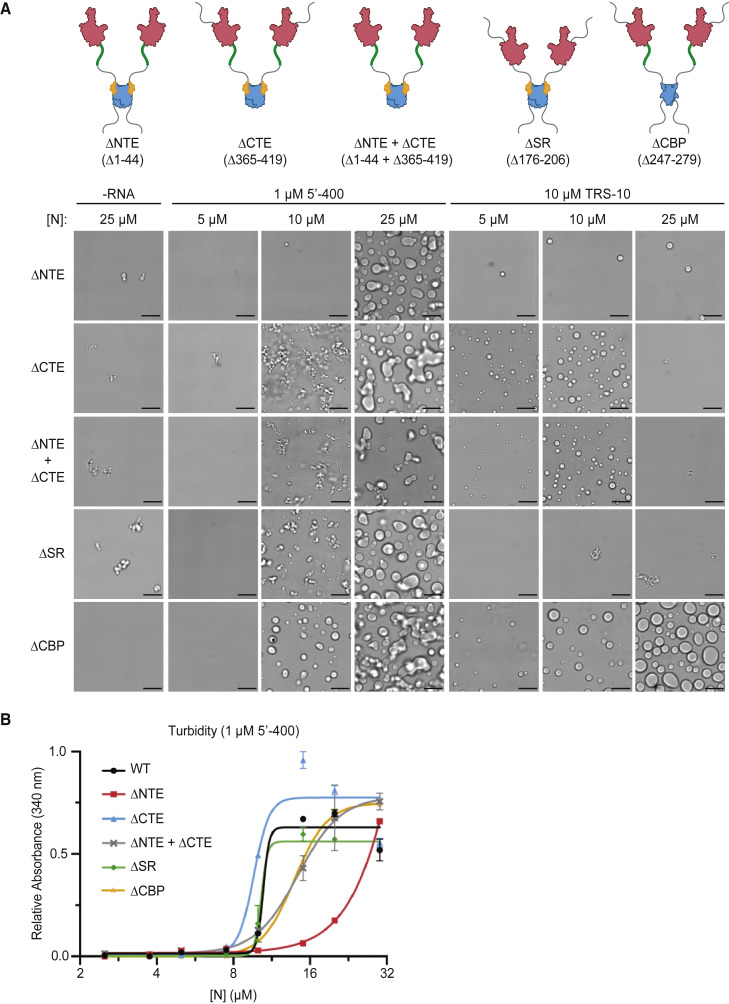
We next explored the roles of N protein disordered regions. We analyzed mutant proteins lacking the following regions (Figure 2 A; Figure S1): (1) the 44-aa N-terminal extension (NTE), a poorly conserved prion-like sequence with a basic cluster that contributes to RNA binding (Chang et al., 2009); (2) the 31-aa SR region, a basic segment implicated in RNA binding, oligomerization (Luo et al., 2005), and phosphorylation (Bouhaddou et al., 2020; Davidson et al., 2020; Fung and Liu, 2018; Klann et al., 2020; Peng et al., 2008; Stohlman et al., 1983; Surjit et al., 2005; White et al., 2007; Wu et al., 2014; Wu et al., 2009); (3) the 55-aa C-terminal extension (CTE), implicated in oligomerization (Hurst et al., 2009; Luo et al., 2006; Luo et al., 2005; Ye et al., 2020); and (4) the CTD basic patch (CBP), a 33-aa basic region that forms the RNA-binding groove on the surface of the CTD (Chen et al., 2007; Takeda et al., 2008; Ye et al., 2020), which can be deleted without affecting CTD dimer structure (Yu et al., 2006).
Figure 2.
Disordered Regions Modulate N Protein Condensate Formation
(A) Top: schematics of N protein deletion mutants. Bottom: N protein condensates observed in the presence of 5′-400 RNA or TRS-10 RNA after a 30-min incubation at room temperature. Images are representative of multiple independent experiments. Scale bar, 10 μm.
(B) Absorbance at 340 nm was used to quantify the turbidity of N protein mixtures after a 15-min incubation at room temperature with 1 μM 5′-400. Data points indicate mean ± SEM of duplicates; representative of two independent experiments.
When combined with viral 5′-400 RNA, none of the deletions completely prevented droplet formation at high protein concentrations (Figure 2A), indicating that no single disordered segment is essential for the interactions that mediate droplet formation. CTE deletion stimulated the formation of abundant filaments, suggesting that this region normally inhibits certain interactions. Deletion of the NTE or CBP abolished filaments. Droplets were also observed after deletion of both the NTE and CTE, showing that the central regions are sufficient for droplet formation. Turbidity analyses (Figure 2B) showed that full-length N protein structures increased abruptly between 5 and 10 μM, supporting a cooperative mechanism of oligomer assembly. Consistent with the microscopy analyses, CTE deletion reduced the saturating concentration and NTE deletion increased it, further supporting the negative and positive roles, respectively, of these regions. Deletion of both the NTE and CTE resulted in an intermediate phenotype (Figure 2B).
Further insights arose in studies of deletion mutants and the 10-nt TRS RNA (Figure 2A). By minimizing the contribution of multivalent RNA binding, these studies illuminated critical protein-protein interactions that contribute to condensate formation. As in the experiments with long RNA, CTE deletion enhanced droplet formation, NTE deletion inhibited it, and the double deletion had little effect, pointing to these regions as opposing but nonessential modulators of protein-protein interactions. As in the wild-type protein (Figure 1B), a molar excess of N protein suppressed droplet formation by TRS RNA in most mutants; only the CBP deletion caused abundant droplets when the protein was in excess of RNA, suggesting that the CBP is responsible for the reduced droplet formation seen with high N protein concentrations. In contrast to results with long RNA, deletion of the SR region abolished TRS-mediated condensates (Figure 2A). TRS binding to the NTD is known to be enhanced by the basic SR region, but SR deletion has only a moderate impact on affinity (Grossoehme et al., 2009). At the RNA concentration used in our experiments, it is unlikely that SR deletion abolished RNA binding. We therefore suspect that the SR region, perhaps in association with part of the RNA, is required for TRS-dependent droplet formation because it mediates a weak interaction with another N protein (Luo et al., 2005).
Phosphorylation Promotes More Liquid-like N Protein Condensates
N protein phosphorylation depends on a poorly understood collaboration between multiple kinases (Peng et al., 2008; Surjit et al., 2005; Wu et al., 2014; Wu et al., 2009). One important player is the abundant cytoplasmic kinase GSK-3, which generally phosphorylates serines or threonines four residues upstream of pre-phosphorylated “priming” sites (Beurel et al., 2015). Studies of the N protein of SARS-CoV (Wu et al., 2009) support the presence of two priming sites, P1 and P2 (Figure 3 A), where phosphorylation initiates a series of GSK-3-mediated phosphorylation events, each primed by the previous site, resulting in a high density of up to ten phosphates. The kinases responsible for priming phosphorylation are not known, but the P2 site (S206 in SARS-CoV-2) is a strong consensus sequence (S/T-P-x-K/R) for Cdk1, a major cell cycle kinase (Morgan, 1997; Surjit et al., 2005).
Figure 3.
Phosphorylation Modulates N Protein Condensate Properties
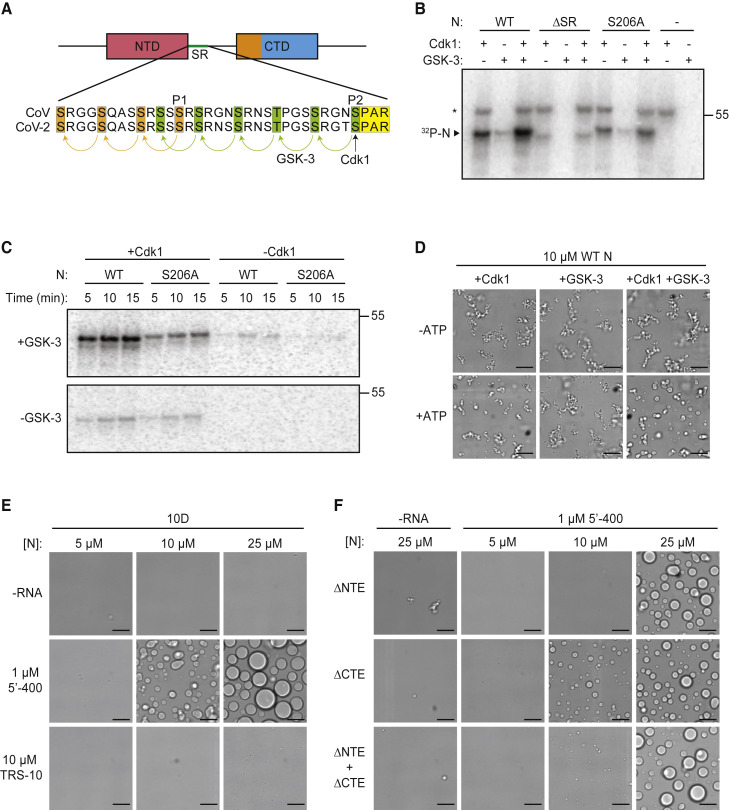
(A) Sequences of the SR regions of SARS-CoV N protein (aa 177–210) and SARS-CoV-2 N protein (aa 176–209). Proposed priming sites (P1 and P2) for GSK-3 are indicated (Wu et al., 2009). P2 (S206 in SARS-CoV-2) is a Cdk consensus site (yellow) where phosphorylation is thought to prime sequential phosphorylation (arrows) of five upstream sites (green) by GSK-3. P1 phosphorylation by an unknown kinase primes phosphorylation at three upstream sites (orange).
(B) The indicated N protein variants were incubated for 30 min with Cdk1-cyclin B1 and/or GSK-3 and radiolabeled ATP, and reaction products were analyzed by SDS-PAGE and autoradiography. Radiolabeled N protein is indicated. Asterisk indicates cyclin B1 autophosphorylation. Molecular weight marker shown on the right (kDa).
(C) N protein was incubated overnight with unlabeled ATP and Cdk1-cyclin B1 (left lanes) or no kinase (right lanes), desalted, and incubated with or without GSK-3 and radiolabeled ATP. Reaction products were analyzed by SDS-PAGE and autoradiography. Molecular weight marker shown on the right (kDa).
(D) 10 μM N protein was incubated 2 h with Cdk1-cyclin B1 and GSK-3 in the presence or absence of ATP, dialyzed into droplet buffer overnight, and mixed with 1 μM 5′-400 RNA. After 30 min, N protein condensates were analyzed by light microscopy.
(E) Images of N protein 10D mutant following 30-min incubation with or without 1 μM 5′-400 or 10 μM TRS-10 RNA.
(F) Images of 10D mutants with the indicated deletions, incubated with or without 1 μM 5′-400 RNA.
(B–F) All results are representative of multiple independent experiments; scale bar, 10 μm.
To produce phosphorylated N protein, we first tested the possibility that Cdk1 primes the protein for subsequent phosphorylation by GSK-3. We found that Cdk1-cyclin B1 phosphorylated N protein in the SR region and mutation of S206 reduced Cdk1-dependent phosphorylation (Figure 3B). Phosphorylation might also occur at T198, a nearby Cdk1 consensus site. A combination of Cdk1 and GSK-3 enhanced phosphorylation. Clear evidence for priming by Cdk1 was obtained by extensive unlabeled phosphorylation by Cdk1, followed by analysis of radiolabeled phosphorylation with GSK-3 (Figure 3C).
Phosphorylation of N protein with a combination of Cdk1 and GSK-3 reduced filamentous structures and promoted the formation of more spherical droplets (Figure 3D). GSK-3 alone had no effect, whereas Cdk1 alone promoted droplets to a small extent. Phosphorylation in the SR region therefore appears to promote the formation of more liquid-like condensates.
We explored the role of phosphorylation in depth with studies of a phosphomimetic mutant in which the ten serines and threonines in the SR region were replaced with aspartate (the 10D mutant). When combined with the 5′-400 viral RNA, the 10D protein rapidly formed condensates with a spherical droplet morphology that was clearly distinct from the filamentous structures of the wild-type protein (Figure 3E). The droplets seen with the 10D mutant were more spherical and uniform than those observed following phosphorylation in vitro (Figure 3D), presumably because phosphorylation at all sites is unlikely to be achieved in vitro. All three viral RNAs were effective in driving droplet formation, although N-1260 appeared to reduce the saturating concentration (Figure S4A). NTE deletion in the 10D protein reduced droplet formation, showing once again the positive role of this region (Figure 3F).
Importantly, the 10D mutation had a greater impact on condensate morphology (Figure 3E) than an SR deletion (Figure 2A), indicating that phosphorylation does not simply block SR function but might also interfere with other interactions. One plausible possibility, for example, is that the abundant negative charge of the phosphorylated SR region interacts intramolecularly with one or more of the positively charged patches on the adjacent NTD or CTD, thereby interfering with multivalent RNA-protein interactions.
Droplet formation by TRS RNA was abolished in the 10D mutant (Figure 3E), just as we observed with TRS RNA and the SR deletion (Figure 2A). These results support the notion that phosphorylation blocks weak protein-protein interactions mediated by the SR region.
Phosphorylated N protein is thought to be localized at the RTC (Stohlman et al., 1983). The N protein of mouse hepatitis virus (MHV) is known to interact directly with the N-terminal Ubl1 domain of Nsp3, a large transmembrane protein localized to RTC membranes (Cong et al., 2020; Hurst et al., 2013; Hurst et al., 2010; Keane and Giedroc, 2013). We found that a GFP-tagged Ubl1 domain of SARS-CoV-2 Nsp3 partitioned into N protein droplets and filamentous structures (Figure S4B), providing a potential mechanism for association of the RTC with N protein condensates.
To gain a better understanding of the properties of N protein structures, we analyzed the fusion dynamics of different structures over time. During a short (90 s) time course, the filamentous structures of 10 μM wild-type protein remained immobile and did not fuse, while the spherical droplets of the 10D mutant were highly dynamic and fused rapidly (Figure 4 A). We also found that the droplet-like structures at higher concentrations of unmodified protein displayed relatively slow fusion activity compared to the droplets of phosphorylated N protein (Figure 4B). These results suggest that unmodified protein forms gel-like condensates that are relatively rigid, while phosphorylated N protein forms condensates that behave more like liquid droplets.
Figure 4.
Phosphorylation of N Protein Promotes Liquid-like Behavior
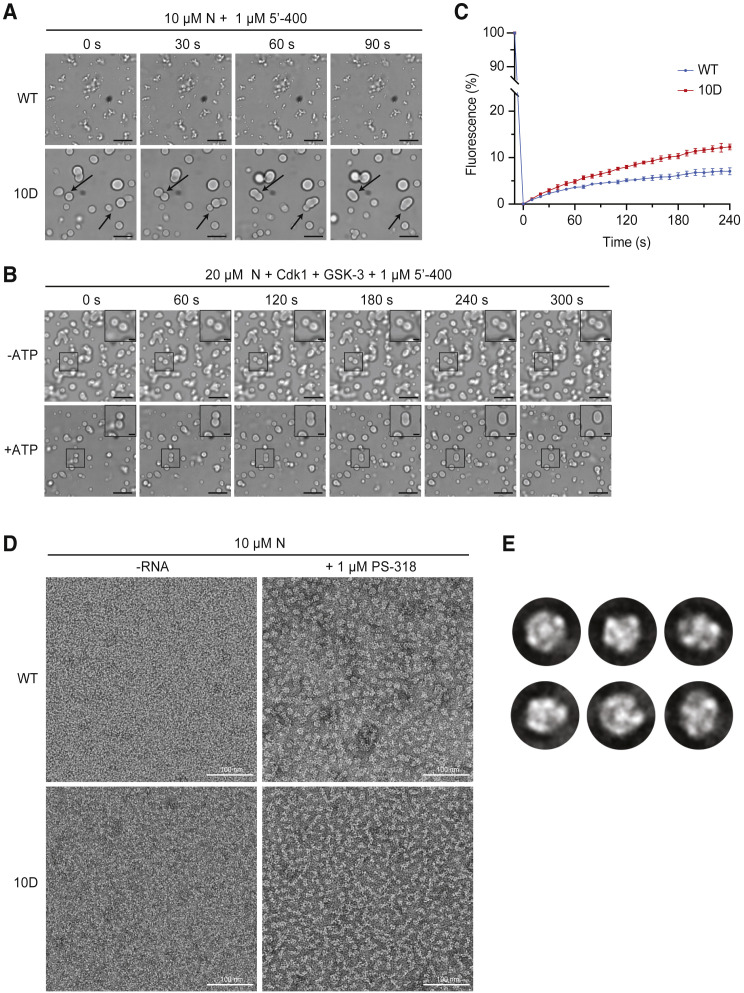
(A) N protein (10 μM wild-type [WT] or 10D) was mixed with 1 μM 5′-400 RNA for 20 min, and images were taken at 30-s intervals. Arrows indicate droplet fusion events in the 10D mutant. No fusion events were observed in WT structures.
(B) 20 μM N protein was phosphorylated with Cdk1-cyclin B1 and GSK-3 as in Figure 3D, incubated with 1 μM 5′-400 RNA for 20 min, and imaged every 60 s. Representative droplet fusion events are boxed with higher magnification images in insets at upper right. Images are representative of multiple independent experiments; scale bar, 10 μm (2 μm for insets).
(C) FRAP analysis of droplets formed with 20 μM dye-labeled N protein (WT or 10D) and 1 μM 5′-400 RNA. Following 30 s of bleaching, droplet fluorescence was measured starting at time zero. Data points indicate mean fluorescence intensity as a percentage of pre-bleaching intensity (mean ± SEM, n = 2 for WT and n = 3 for 10D). Note the break in the y axis to allow better viewing of recovery data. Results are representative of two independent experiments. See Figure S4C for images.
(D) 10 μM N protein (WT or 10D) was incubated without or with 1 μM PS-318 RNA for 15 min prior to analysis by negative-stain electron microscopy. Images are representative of three independent experiments. Scale bar, 100 nm.
(E) 2D class averages of particles from the EM analysis of WT N protein and RNA in (D). Particle selection was not possible with the nonuniform structures formed by the 10D mutant.
We analyzed condensate properties further by labeling N protein with a fluorescent dye, incubating 20 μM N protein with 5′-400 RNA to form droplets, and measuring fluorescence recovery after photobleaching (FRAP). Wild-type N protein droplets recovered slowly (about 7% over 4 min), and recovery was most apparent at the edges of the droplets (Figures 4C and S4C). 10D mutant droplets recovered more uniformly and about 2-fold more rapidly (12% over 4 min), suggesting that phosphorylation generates a more dynamic condensate but also showing that both wild-type and 10D condensates are less dynamic in vitro than many previously described condensates.
We used negative-stain electron microscopy to further analyze mixtures of 10 μM N protein and 1 μM viral RNA (Figure 4D). Unmodified wild-type protein and PS-318 RNA formed particles of ∼15 nm diameter, and two-dimensional classification of these particles revealed a uniform size and architecture (Figure 4E). Similar particles were observed with 5′-400 RNA (Figure S4D). Thus, the gel-like filamentous condensates of the unmodified protein—and possibly the nucleocapsid—are likely to be assembled on a foundation of discrete structural building blocks. In contrast, the mixture of 10D mutant and RNA formed nonuniform, poorly defined chains (Figure 4D), suggesting that phosphorylation disrupts the structural units of the unmodified protein to create a more diffuse, liquid-like condensate.
Discussion
We conclude that the N protein of SARS-CoV-2, together with viral RNA, assembles into multiple structural forms that depend on a complex blend of intramolecular and intermolecular interactions. The more rigid filamentous condensates of the unmodified protein most likely depend on high-avidity interactions mediated by multivalent RNA-protein, RNA-RNA, and protein-protein interactions. The latter might include prion-like interactions between NTEs, binding of the SR region to the CTD (Luo et al., 2005), or helical CTD polymers that depend on the CBP (Chen et al., 2007; Gui et al., 2017). Long RNAs augment these protein-protein interactions by interacting with the numerous RNA-binding sites on the protein. Our results also support a model in which phosphorylation of the SR region blocks SR-mediated protein-protein interactions and interferes intramolecularly with RNA binding at other sites, resulting in a loss of affinity that generates a more dynamic liquid droplet.
The different forms of N protein oligomers seem well suited for its two major functions. In the nucleocapsid, where extremely compact RNA packaging is the goal, the organized structures of the unmodified protein could represent an underlying structural framework that is supplemented by liquid-like condensation—much like chromosome packaging depends on underlying nucleosome structure and the condensate behavior of chromatin proteins (Larson et al., 2017; Larson and Narlikar, 2018). Consistent with this possibility, the ribonucleoprotein particles we observed with unmodified N protein and viral RNA are remarkably similar in size and shape to nucleocapsid particles seen in electron microscopy and tomography studies of partially disrupted MHV nucleocapsids (Gui et al., 2017) and SARS-CoV-2 virions (Klein et al., 2020; Yao et al., 2020).
The more liquid-like behavior of phosphorylated N protein condensates might be particularly important at the RTC. There is abundant evidence linking N protein phosphorylation with localization and function in the RTC. N protein is fully phosphorylated soon after synthesis and rapidly associates with large membrane structures that presumably represent the RTC (Stohlman et al., 1983). N protein is the only viral structural protein that localizes to the RTC (Snijder et al., 2020; V’kovski et al., 2019), and immunoelectron microscopy reveals high concentrations of N protein in electron-dense regions adjacent to the double-membrane vesicles and convoluted membranes of the RTC (Snijder et al., 2020; Stertz et al., 2007; Ulasli et al., 2010). At these sites, N protein co-localizes with the transmembrane viral protein Nsp3, which is known to bind N protein and might therefore anchor N protein oligomers to RTC membranes (Cong et al., 2020; Hurst et al., 2013; Hurst et al., 2010; Keane and Giedroc, 2013; Wolff et al., 2020) (Figure S4B). In light microscopy of infected cells, GFP-tagged N protein forms large cytoplasmic granules that co-localize with Nsp3 at RTCs (Bost et al., 2000; Cong et al., 2020; Stertz et al., 2007; Verheije et al., 2008), and FRAP indicates that N protein is dynamically associated with these granules (Verheije et al., 2010). N protein helps control subgenomic transcription in the RTC, and inhibition of phosphorylation blocks this function (Almazán et al., 2004; Wu et al., 2014; Zúñiga et al., 2010). With these many lines of evidence in mind, our work points to the possibility that a condensate of phosphorylated N protein and loosely bound RNA, linked to RTC membranes by Nsp3, provides a compartment to concentrate and protect the viral replication and transcription machinery. Similar mechanisms are likely to exist in negative-sense RNA viruses, where replication is focused in dynamic biomolecular condensates (Heinrich et al., 2018; Nevers et al., 2020; Nikolic et al., 2017; Zhou et al., 2019). In measles virus, these condensates have also been implicated in nucleocapsid assembly (Guseva et al., 2020).
Others have also observed recently that the N protein of SARS-CoV-2 forms RNA-dependent biomolecular condensates in vitro (Chen et al., 2020; Cubuk et al., 2020; Iserman et al., 2020; Jack et al., 2020; Lu et al., 2020; Perdikari et al., 2020; Savastano et al., 2020). When overexpressed at high levels in uninfected cells, the N proteins of MHV (Verheije et al., 2008), SARS-CoV (Peng et al., 2008), and SARS-CoV-2 (Iserman et al., 2020; Lu et al., 2020) form cytoplasmic granules. In the case of SARS-CoV-2 N protein, these granules display the fusion and rapid turnover behaviors of dynamic condensates (Iserman et al., 2020; Lu et al., 2020). Most importantly, as mentioned above, the granules of N protein that form at the RTC in infected cells also display rapid turnover (Verheije et al., 2010).
FRAP reveals that N protein granules in the cell turn over much more rapidly than N protein condensates in vitro (Figure 4C) (Iserman et al., 2020; Lu et al., 2020; Verheije et al., 2010). Phosphorylation in vivo does not entirely explain this discrepancy as we found that the 10D mutant still displays relatively slow turnover. We speculate that N protein dynamics in the cell, and particularly in the infected cell, are further governed by cellular and viral proteins and RNA.
There is some evidence to suggest a link between N protein and stress granules, the cytoplasmic mRNA-protein condensates that form in response to translation arrest and contribute to the response to some viral infections (Cascarina and Ross, 2020; McCormick and Khaperskyy, 2017; Protter and Parker, 2016; Tsai and Lloyd, 2014). In uninfected cells, overexpressed N protein of SARS-CoV (Peng et al., 2008) or SARS-CoV-2 (Savastano et al., 2020) associates with stress granules, and N protein associates in vitro with condensates of stress granule proteins (Perdikari et al., 2020). Recent proteomic analyses indicate that the N protein of SARS-CoV-2, when expressed in uninfected cells, associates with proteins found in stress granules (Gordon et al., 2020; Li et al., 2020; Samavarchi-Tehrani et al., 2020; Stukalov et al., 2020) and appears to suppress stress granule formation (Nabeel-Shah et al., 2020; Samavarchi-Tehrani et al., 2020). Studies of infected cells will be required, however, to determine if stress granules form during SARS-CoV-2 infection and whether N protein has any role in their regulation.
Consistent with our results, N protein granules in MHV-infected cells display dramatic changes in morphology following chemical inhibition of GSK-3 kinase activity (Wu et al., 2014). Most importantly, GSK-3 inhibition disrupts MHV genome processing and reduces the production of virions by MHV- or SARS-CoV-infected cells (Wu et al., 2014; Wu et al., 2009). Cdk inhibitors inhibit virus production by SARS-CoV-2-infected cells (Bouhaddou et al., 2020). Viral replication is also reduced by inhibition of the kinases SRPK1 and 2, which have also been implicated in N protein phosphorylation (Heaton et al., 2020; Nikolakaki and Giannakouros, 2020; Peng et al., 2008). Thus, chemical inhibitors of kinases that phosphorylate the N protein have the potential to serve as antiviral therapies in the early stages of COVID-19.
Limitations
Our studies focused on the behavior of purified N protein in vitro when mixed with fragments of the viral RNA genome. As in all studies of purified components, it is not yet clear that the interesting behaviors we observe are relevant to N protein function in the infected cell, where numerous host and viral proteins and RNAs are likely to comingle with N protein at the RTC and at viral packaging sites. It is also possible that phosphoregulation of the N protein in the cell is complex and that specific sites and kinases play more nuanced roles in the control of N protein interactions with itself and other components. The resolution of these questions will require detailed microscopic and biochemical analyses of N protein behavior, viral genome transcription, and viral packaging in cells infected with wild-type SARS-CoV-2 or with viruses bearing mutations in phosphorylation sites or other sites that modulate condensate behavior.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| One Shot™ BL21 Star™ (DE3) | Thermo Fisher | Cat # C601003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phenylmethanesulfonyl fluoride (PMSF) | Sigma | Cat # P7626 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat # 4693159001 |
| 10X TBE | BioRad | Cat # 1610733 |
| Ulp1 (aa 406-622) | This study | N/A |
| IPTG | Grow cells | Cat # MESP-2002 |
| Imidazole | Sigma | Cat # 1202 |
| Human GSK-3β | Promega | Cat # V1991 |
| Human Cdk1-cyclin B1 | This study | N/A |
| SARS-CoV-2 N protein constructs | This study | N/A |
| SARS-CoV-2 Nsp3 Ubl1 domain (aa 1-110)-GFP | This study | N/A |
| 32P-γ-ATP | Perkin Elmer | Cat # BLU502A250UC |
| Glass bottom 384-well plate | Greiner Bio | Cat # 781892 |
| mPEG-Saline | Laysan Bio | Item # MPEG-SIL-5000 |
| ATP | Sigma | Cat # A26209 |
| Uranyl Formate | VWR | Cat # 101410-834 |
| Glider Hexagonal Grid 400 mesh, copper | Electron microscopy science | Cat # G400H-Cu |
| 10% TBE-Urea gel | Invitrogen | Cat # EC68755 |
| 10% Mini-PROTEAN® TGX™ Precast Protein Gels | BioRad | Cat # 4561036 |
| Alexa Fluor™ 488 NHS Ester | Invitrogen | Cat # A20000 |
| SYBR Gold | Invitrogen | Cat # S11494 |
| Desthiobiotin | Sigma | Cat # D1411 |
| Phosphocreatine | Sigma | Cat # 10621714001 |
| Creatine Kinase | Sigma | Cat # C3755 |
| Critical Commercial Assays | ||
| HiScribe™ T7 High Yield RNA Synthesis Kit | NEB | Cat # E2040S |
| Superdex S200 10/300 GL | GE | Cat # 17-5175-01 |
| Amicon Ultra-0.5 Centrifugal Filter Unit – 30kDa | Millipore | Cat # UFC503096 |
| Amicon Ultra-15 Centrifugal Filter Unit – 30kDa | Millipore | Cat # UFC903024 |
| Ni-NTA Agarose | QIAGEN | Cat # 30230 |
| Zeba™ Spin Desalting Columns, 7K MWCO, 0.5 mL | Thermo | Cat # 89882 |
| Dialysis membrane | Spectrum Labs | Cat # 132676 |
| RNA Clean & Concentrator | Zymo Research | Cat # 11-353 |
| HisTRAP column | Cytiva | Cat # 17524701 |
| StrepTRAP column | Cytiva | Cat # 28907546 |
| Experimental Models: Cell Lines | ||
| Sf9 | Thermo | Cat # 11496015 |
| Recombinant DNA | ||
| pLIB: Cdk1 (untagged) | This study | N/A |
| pLIB: 6xHis-Myc-Cyclin B1 | This study | N/A |
| pET28a: 6xHIS-tev-Ulp1 (406-622) | This study | N/A |
| pET28a: 6xHIS-Smt3-N | This study | N/A |
| pET28a: 6xHIS-Smt3-N ΔSR | This study | N/A |
| pET28a: 6xHIS-Smt3-N ΔNTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N ΔCTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N ΔCBP | This study | N/A |
| pET28a: 6xHIS-Smt3-N ΔNTE+ΔCTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N 10D | This study | N/A |
| pET28a: 6xHIS-Smt3-N 10D ΔNTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N 10D ΔCTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N 10D ΔNTE+ΔCTE | This study | N/A |
| pET28a: 6xHIS-Smt3-N S206A | This study | N/A |
| pET28a: Nsp3 (1-110)-GFP-2xStrep | This study | N/A |
| Constructs for RNAs used in this study (Table S1) | This study | N/A |
| Software and Algorithms | ||
| Prism 8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | Open source | https://imagej.net/Welcome |
| FIJI | Open source | https://imagej.net/Fiji |
| Relion-3 | (Zivanov et al., 2018) | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Download_%26_install |
| Other | ||
| Uncropped microscopy images are available from Mendeley Data: https://data.mendeley.com/datasets/88xpyp9prs/2 | N/A | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Morgan (david.morgan@ucsf.edu).
Materials Availability
Materials such as plasmid constructs will be available without further restrictions upon request to the Lead Contact (david.morgan@ucsf.edu).
Data and Code Availability
Uncropped microscopy images are available from Mendeley Data: https://data.mendeley.com/datasets/88xpyp9prs/2.
Experimental Model and Subject Details
All recombinant proteins were expressed in One Shot BL21 Star (DE3) chemically competent E. coli (Thermo). The genotype is: F- ompT hsdSB (rB -, mB -) galdcmrne131 (DE3).
Method Details
Plasmid construction and RNA preparation
All expression vectors were constructed by Gibson assembly and confirmed by sequencing. Codon-optimized DNAs encoding SARS-CoV-2 N protein (aa 1-419) and Nsp3 Ubl1 domain (aa 819-920 of polyprotein 1a) were ordered as gBlocks from Integrated DNA Technologies (IDT) and cloned into pET28a expression vectors. N protein mutants were constructed by site-directed mutagenesis and Gibson assembly. A 6xHis-SUMO tag was added to the N terminus of all N protein constructs. Nsp3 Ubl1 domain was engineered with a C-terminal GFP-2xStrep tag. Codon-optimized DNAs encoding human Cdk1 (untagged) and Cyclin B1 (N-terminal 6xHis-4xMyc tag) were ordered as gBlocks from IDT and cloned individually into the baculovirus expression vector pLIB for construction of recombinant baculoviruses (Weissmann et al., 2016). Human GSK-3β was purchased from Promega (V1991).
Sequences of the three viral RNAs and firefly luciferase RNA are provided in Table S1. Templates for transcription in vitro of 5′-400 RNA and PS-318 RNA were ordered as gBlocks from IDT and PCR-amplified with a 5′ primer carrying a T7 promoter sequence. The template for N-1260 was PCR-amplified with a 5′ primer containing a T7 promoter from a PET28a vector carrying PCR-amplified ORF DNA from the 2019-nCoV_N positive control plasmid from IDT. The N-1260 RNA includes 152 nucleotides from the pET28a plasmid backbone. All RNA synthesis was performed using the HiScribe T7 High Yield RNA synthesis kit (NEB E2040S) according to the manufacturer’s protocol. Luc-1710 RNA was included as a positive control template in the RNA synthesis kit. The SARS-CoV-2 TRS RNA (UCUAAACGAA, tagged with FAM fluorescent dye) was ordered from IDT.
Protein purification
N protein vectors were transformed into E. coli BL21 Star (Thermo #C601003) for expression. Freshly transformed cells were grown in TB-Kanamycin (50 μg/mL) to OD 0.4 at 37°C. The temperature was lowered to 16°C until the cells reached a density of 0.8. Protein expression was then induced with 0.4 mM IPTG for 16 h. Harvested cells were washed with PBS, snap frozen in LN2, and stored at −80°C until lysis.
To remove contaminating nucleic acid, N protein was purified under denaturing conditions as previously described (Peng et al., 2008; Wang et al., 2004). Frozen cell pellets were thawed and resuspended in buffer A (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 20 mM imidazole, 6 M urea) and lysed by sonication on ice. Lysed cells were centrifuged at 30,000 rpm for 30 min at 4°C to remove cell debris. Clarified lysate was added to Ni-NTA agarose beads (QIAGEN) and incubated for 45 min at 4°C. Ni-NTA beads were then washed 3 times with 10 bed volumes of buffer A, and N protein was eluted with 3 bed volumes buffer B (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 250 mM imidazole, 6 M urea). The eluate was concentrated to ∼1 mL using centrifugal concentrators (30 kDa cutoff, Amicon) and renatured by overnight dialysis against 2 l buffer C (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol) at 4°C. 100 μg recombinant Ulp1 catalytic domain (expressed in and purified from bacteria) was added to renatured protein for 10 min on ice to cleave the 6xHis-SUMO tag. Mutants lacking the NTE were not cleaved as efficiently by Ulp1, and complete cleavage required incubation with Ulp1 at 25°C for 4 h. Cleaved protein was then centrifuged at 15,000 rpm for 10 min and injected onto a Superdex 200 10/300 size exclusion column equilibrated in buffer C. Peak fractions were pooled, frozen in LN2, and stored at −80°C.
In early experiments, N protein was purified under native conditions. Frozen cell pellets were thawed and resuspended in buffer D (50 mM HEPES pH 7.5, 500 mM NaCl, 10% glycerol, 20 mM imidazole) supplemented with benzonase, cOmplete EDTA-free protease inhibitor cocktail (Roche), and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed by sonication on ice and centrifuged at 30,000 rpm for 30 min at 4°C. Clarified lysate was added to Ni-NTA agarose beads and incubated for 45 min at 4°C. Ni-NTA beads were then washed 3 times with 10 bed volumes buffer D, and N protein was eluted with 3 bed volumes buffer D + 500 mM imidazole. The eluate was concentrated to ∼1 mL using centrifugal concentrators (30 kDa cutoff, Amicon) and injected onto a Superdex 200 10/300 size exclusion column. Peak fractions were pooled, snap frozen in LN2, and stored at −80°C. Protein concentration was measured by nanodrop, and a major A260 peak was observed. The A260 peak was insensitive to treatment with DNase I, RNase A, RNase H, or benzonase. Additionally, small RNA species were observed on native TBE gels stained with SYBR gold.
Nsp3 Ubl1-GFP was expressed in E. coli as described above. Frozen cell pellets were thawed and resuspended in buffer E (50 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM DTT) supplemented with benzonase, cOmplete EDTA-free protease inhibitor cocktail (Roche), and 1 mM PMSF. Cells were lysed by sonication on ice and centrifuged at 30,000 rpm for 30 min at 4°C. Clarified lysate was then added to a 5 mL StrepTrap HP prepacked column (Cytiva). The column was washed with 10 column volumes buffer E and eluted with 4 column volumes buffer E supplemented with 2.5 mM desthiobiotin. Peak fractions were pooled, snap frozen in LN2, and stored at −80°C.
Cdk1-cyclin B1 complexes were prepared as follows. Two 666 mL cultures of SF-900 cells (1.6106 cells/mL) were infected separately with Cdk1 or cyclin B1 baculovirus and harvested after 48 h. The two cell pellets were frozen in LN2. Frozen pellets were thawed and each resuspended in 20 mL lysis buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol, 20 mM imidazole, benzonase, and cOmplete EDTA-free protease inhibitor cocktail). Cells were lysed by sonication. To generate an active Cdk1-cyclin B1 complex phosphorylated by Cdk-activating kinase in the lysate (Desai et al., 1992), the two lysates were combined, brought to 5 mM ATP and 10 mM MgCl2, and incubated at room temperature for 20 min. The combined lysates were centrifuged at 55,000 rpm for 45 min at 4°C. The supernatant was filtered and passed over a HisTRAP nickel affinity column, washed with wash buffer (50 mM HEPES pH 7.5, 300 mM NaCl, 10% glycerol) and eluted with the same buffer plus 200 mM imidazole. Peak fractions were pooled, concentrated to 0.75 ml, and injected on an S200 size exclusion column in wash buffer. Peak fractions containing the Cdk1-cyclin B1 complex were pooled, concentrated to 1.5 mg/mL, and snap frozen in LN2.
Light microscopy
Glass was prepared as described previously (Keenen et al., 2018). Individual wells in a 384-well glass bottom plate (Greiner #781892) were incubated with 2% Hellmanex detergent for 1 h. Wells were then washed 3 times with 100 μL ddH2O. 1 M NaOH was added to the glass for 30 min, followed by washing 3 times with 100 μL ddH2O. The glass was dried, and 20 mg/mL PEG silane dissolved in 95% EtOH was added to individual wells and incubated overnight (∼16 h). The glass was then washed 3 times with 100 μL ddH2O and dried before sample addition.
The day prior to imaging, protein was thawed and dialyzed against droplet buffer (25 mM HEPES pH 7.5, 70 mM KCl) overnight at 4°C. Protein concentration was quantified by nanodrop. Reactions containing protein and RNA were combined and mixed immediately before adding to individual wells in the PEG-treated 384-well plate. All reactions were incubated for 30 min at room temperature, unless otherwise indicated, before imaging on a Zeiss Axiovert 200M microscope with a 40x oil objective.
Turbidity analysis
Protein was dialyzed into droplet buffer the night before performing turbidity measurements, as stated above. Protein was then serially diluted into room temperature droplet buffer. After dilution, RNA was added and mixtures were incubated at room temperature for 15 min before measuring absorbance at 340 nm on a Spectramax M5 plate reader.
Kinase reactions
1 μM N protein was incubated with either 0.14 μM GSK-3β, 0.91 μM Cdk1-Cyclin B1, or both, in a 20 μL reaction mixture containing 10 mM MgCl2, 1 mM DTT, 500 μM ATP, 25 mM HEPES pH 7.8, 50 mM NaCl, and 0.001 mCi/mL 32P-γ-ATP. After incubation at 30°C for 30 min, reactions were quenched with 10 μL SDS loading buffer for analysis by SDS-PAGE and visualization with a Phosphorimager. For the priming experiment in Figure 3C, 19 μM of each N protein variant was incubated with 2.5 μM Cdk1-Cyclin B1 overnight at 25°C in kinase buffer A (20 mM HEPES pH 7.4, 300 mM NaCl, 20 mM MgCl2, 1 mM DTT, 1 mM ATP). Reactions were desalted into kinase buffer B (20 mM HEPES pH 7.4, 20 mM MgCl2, 1 mM DTT, 500 μM ATP). 10 μM of desalted N protein was incubated with 0.17 μM GSK-3β and 0.05 μCi 32P-γ-ATP. After various times, reactions were quenched with SDS loading buffer for analysis by SDS-PAGE and visualization with a Phosphorimager.
Phosphorylated protein for droplet analysis was prepared in reactions containing 25 μM N protein, 1 μM Cdk1-cyclin B1, 0.2 μM GSK-3β, 2 mM ATP, 20 mM MgCl2, 15 mM HEPES pH 7.5, 300 mM NaCl, 5% glycerol, 1 mM DTT, 8 mM phosphocreatine (Sigma 10621714001), and 0.016 mg/mL creatine kinase (Sigma C3755). The reaction was incubated for 2 h at 30°C and then dialyzed overnight into droplet buffer at 4°C. RNA addition and droplet visualization were carried out as described above.
Fluorescence recovery after photobleaching (FRAP)
Purified N protein (wild type or 10D) was labeled by incubation with 3:1 Alexa Fluor 488 NHS Ester (Invitrogen A20000) at 30°C for 45 min. N protein was then purified by size exclusion chromatography, concentrated, and stored at −80°C until use. For FRAP experiments, glass coverslips and slides were cleaned and functionalized with polyethylene glycol as previously described (Jain et al., 2012), and chambers were constructed with double-sided tape. 20 μM N protein, diluted to 5% labeled N with unlabeled protein, was mixed with 1 μM 5′-400 RNA and immediately added to preassembled glass chambers. Fluorescent droplets were visualized by Total Internal Reflection Fluorescence on a DeltaVision OMX SR microscope with a 60x objective (immersion oil refractive index: 1.516). Images were acquired at a 1 s frame rate for 3 frames, bleached 30 s at 5% absolute transmission at 405 nm, and imaged immediately thereafter. All experiments were performed on droplets of similar size (2.3-2.6 μm), 15-30 min after RNA addition.
Electron microscopy
N protein (10 μM) and RNA (1 μM PS-318 or 5′-400) were mixed in 10 μL droplet buffer and incubated 15 min at room temperature. 3.5 μL of this solution was adsorbed onto glow-discharged (PELCO EasiGlow, 15 mA, 0.39 mBar, 30 s) carbon-coated grids (200-400 mesh copper, Quantifoil) for 1 min at room temperature. Sample was blotted off, stained and blotted 5 times with 0.75% uranyl formate, and allowed to air dry. Negative stain images were collected with a Tecnai T12 microscope (FEI) with a LaB6 filament, operated at 120 kV, and a Gatan Ultrascan CCD camera (final pixel size 2.21 Å). Contrast Transfer Function (CTF) estimation was performed with CTFFIND4 (Rohou and Grigorieff, 2015). Automated particle picking, CTF-correction, and two-dimensional averaging and classification were performed in RELION-3 (Zivanov et al., 2018).
Quantification and Statistical Analysis
Turbidity Analysis
Turbidity was measured at 340 nm on a Spectramax M5 plate reader, as described in Method Details. Turbidity measurements were performed in duplicate in two independent experiments. Results were normalized to the maximum absorbance signal and analyzed with GraphPad Prism.
Fluorescence recovery after photobleaching
Image analysis was performed with ImageJ. Fluorescence intensity for the whole droplet was calculated, and background intensity subtracted. Intensity before bleaching was normalized to 100% and intensity after bleaching was normalized to 0%. Data were analyzed with GraphPad Prism.
Acknowledgments
We thank Geeta Narlikar, Sy Redding, and Adam Frost for valuable discussions; Madeline Keenen and Emily Wong for biochemical reagents and technical advice; and Delaine Larsen and So Yeon Kim of the UCSF Nikon Imaging Center for assistance with FRAP microscopy. This work was supported by the National Institute of General Medical Sciences, United States (R35-GM118053) and the UCSF Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation, United States.
Author Contributions
C.R.C., J.B.A., C.M.G., C.J.H., N.H., and M.S. contributed to conceptualization, experimental design, and generation of results; C.R.C. prepared proteins and performed microscopy analysis of condensates; J.B.A. and C.M.G. prepared proteins and performed kinase experiments; C.J.H. performed EM analysis; N.H. performed FRAP analysis; M.S. assisted with protein expression and microscopy with guidance from A.D.F.; and D.O.M. provided guidance and wrote the paper with contributions from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.11.025.
Supplemental Information
References
- Alberti S., Gladfelter A., Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán F., Galán C., Enjuanes L. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurel E., Grieco S.F., Jope R.S. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 2000;74:3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascarina S.M., Ross E.D. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020 doi: 10.1096/fj.202001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Chen C.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein--implication for virus ribonucleoprotein packaging. PLoS ONE. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein--forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Cui Y., Han X., Hu W., Sun M., Zhang Y., Wang P.H., Song G., Chen W., Lou J. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. 2020 doi: 10.1038/s41422-020-00408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Kriegenburg F., de Haan C.A.M., Reggiori F. Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci. Rep. 2017;7:5740. doi: 10.1038/s41598-017-06062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., Thiel V., de Haan C.A.M., Reggiori F. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J. Virol. 2020;94:e01925-19. doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. bioRxiv. 2020 doi: 10.1101/2020.06.17.158121. 2020.06.17.158121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- Desai D., Gu Y., Morgan D.O. Activation of human cyclin-dependent kinases in vitro. Mol. Biol. Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd, Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Liu X., Guo D., Zhang Z., Yin C.C., Chen Y., Xiang Y. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell. 2017;8:219–224. doi: 10.1007/s13238-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseva S., Milles S., Jensen M.R., Salvi N., Kleman J.P., Maurin D., Ruigrok R.W.H., Blackledge M. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci. Adv. 2020;6:eaaz7095. doi: 10.1126/sciadv.aaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., Cutts T., Andonov A., Cao J., Booth T.F. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;316:476–483. doi: 10.1016/j.bbrc.2004.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton B.E., Trimarco J.D., Hamele C.E., Harding A.T., Tata A., Zhu X., Tata P.R., Smith C.M., Heaton N.S. SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.14.251207. 2020.08.14.251207. [DOI] [Google Scholar]
- Heinrich B.S., Maliga Z., Stein D.A., Hyman A.A., Whelan S.P.J. Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. MBio. 2018;9:e02290-17. doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H., Chen I.Y., Chang C.K., Huang T.H., Chang M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83:7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013;87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iserman C., Roden C., Boerneke M., Sealfon R., McLaughlin G., Jungreis I., Park C., Boppana A., Fritch E., Hou Y.J. Specific viral RNA drives the SARS CoV-2 nucleocapsid to phase separate. bioRxiv. 2020 doi: 10.1101/2020.06.11.147199. 2020.06.11.147199. [DOI] [Google Scholar]
- Jack A., Ferro L.S., Trnka M.J., Wehri E., Nadgir A., Costa K., Schaletzky J., Yildiz A. SARS CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. bioRxiv. 2020 doi: 10.1101/2020.09.14.295824. 2020.09.14.295824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Liu R., Xiang Y.K., Ha T. Single-molecule pull-down for studying protein interactions. Nat. Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Giedroc D.P. Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J. Virol. 2013;87:3502–3515. doi: 10.1128/JVI.03112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Liu P., Leibowitz J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenen M.M., Larson A.G., Narlikar G.J. Visualization and Quantitation of Phase-Separated Droplet Formation by Human HP1α. Methods Enzymol. 2018;611:51–66. doi: 10.1016/bs.mie.2018.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann K., Bojkova D., Tascher G., Ciesek S., Munch C., Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol. Cell. 2020;80:164–174.e4. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A.G., Narlikar G.J. The Role of Phase Separation in Heterochromatin Formation, Function, and Regulation. Biochemistry. 2018;57:2540–2548. doi: 10.1021/acs.biochem.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Masters P.S. The coronavirus nucleocapsid protein. In: Siddell S.G., editor. The Coronaviridae. Plenum Press; New York: 1995. pp. 141–163. [Google Scholar]
- Li J., Guo M., Tian X., Liu C., Wang X., Yang X., Wu P., Xiao Z., Qu Y., Yin Y. Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med. 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Ye Q., Singh D., Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 Nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. bioRxiv. 2020 doi: 10.1101/2020.07.30.228023. 2020.07.30.228023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Ye F., Sun T., Yue L., Peng S., Chen J., Li G., Du Y., Xie Y., Yang Y. In vitro biochemical and thermodynamic characterization of nucleocapsid protein of SARS. Biophys. Chem. 2004;112:15–25. doi: 10.1016/j.bpc.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Ye F., Chen K., Shen X., Jiang H. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- Luo H., Chen J., Chen K., Shen X., Jiang H. Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Nabeel-Shah N., Lee H., Ahmed N., Marcon E., Farhangmehr S., Pu S., Burke G.L., Ashraf K., Wei H., Zhong G. SARS-CoV-2 nucleocapsid protein attenuates stress granule formation and alters gene expression via direct interaction with host mRNAs. bioRxiv. 2020 doi: 10.1101/2020.10.23.342113. [DOI] [Google Scholar]
- Neuman B.W., Angelini M.M., Buchmeier M.J. Does form meet function in the coronavirus replicative organelle? Trends Microbiol. 2014;22:642–647. doi: 10.1016/j.tim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers Q., Albertini A.A., Lagaudrière-Gesbert C., Gaudin Y. Negri bodies and other virus membrane-less replication compartments. Biochim. Biophys. Acta. Mol. Cell Res. 2020;1867:118831. doi: 10.1016/j.bbamcr.2020.118831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E., Giannakouros T. SR/RS Motifs as Critical Determinants of Coronavirus Life Cycle. Front. Mol. Biosci. 2020;7:219. doi: 10.3389/fmolb.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudrière-Gesbert C., Gaudin Y., Blondel D. Negri bodies are viral factories with properties of liquid organelles. Nat. Commun. 2017;8:58. doi: 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen I., Shewmaker F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019;20:E5501. doi: 10.3390/ijms20215501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS‐CoV‐2 nucleocapsid protein phase‐separates with RNA and with human hnRNPs. EMBO J. 2020 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter D.S.W., Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Abdouni H., Knight J.D.R., Astori A., Samson R., Lin Z.Y., Kim D.K., Knapp J.J., St-Germain J., Go C.D. A SARS-CoV-2 - host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. [DOI] [Google Scholar]
- Savastano A., Ibanez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. bioRxiv. 2020 doi: 10.1101/2020.06.18.160648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Decroly E., Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman S.A., Fleming J.O., Patton C.D., Lai M.M. Synthesis and subcellular localization of the murine coronavirus nucleocapsid protein. Virology. 1983;130:527–532. doi: 10.1016/0042-6822(83)90106-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov A., Girault V., Grass V., Bergant V., Karayel O., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv. 2020 doi: 10.1101/2020.06.17.156455. [DOI] [PubMed] [Google Scholar]
- Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M., Chang C.K., Ikeya T., Güntert P., Chang Y.H., Hsu Y.L., Huang T.H., Kainosho M. Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J. Mol. Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D., Tauber G., Parker R. Mechanisms and Regulation of RNA Condensation in RNP Granule Formation. Trends Biochem. Sci. 2020;45:764–778. doi: 10.1016/j.tibs.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.C., Lloyd R.E. Cytoplasmic RNA Granules and Viral Infection. Annu. Rev. Virol. 2014;1:147–170. doi: 10.1146/annurev-virology-031413-085505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M., Verheije M.H., de Haan C.A., Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 2010;12:844–861. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife. 2019;8:e42037. doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Raaben M., Mari M., Te Lintelo E.G., Reggiori F., van Kuppeveld F.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheije M.H., Hagemeijer M.C., Ulasli M., Reggiori F., Rottier P.J., Masters P.S., de Haan C.A. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 2010;84:11575–11579. doi: 10.1128/JVI.00569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu X., Wang Y., Li B., Zhou H., Yuan G., Fu Y., Luo Y. Low stability of nucleocapsid protein in SARS virus. Biochemistry. 2004;43:11103–11108. doi: 10.1021/bi049194b. [DOI] [PubMed] [Google Scholar]
- Weissmann F., Petzold G., VanderLinden R., Huis In ’t Veld P.J., Brown N.G., Lampert F., Westermann S., Stark H., Schulman B.A., Peters J.M. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc. Natl. Acad. Sci. USA. 2016;113:E2564–E2569. doi: 10.1073/pnas.1604935113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.C., Yi Z., Hogue B.G. Identification of mouse hepatitis coronavirus A59 nucleocapsid protein phosphorylation sites. Virus Res. 2007;126:139–148. doi: 10.1016/j.virusres.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grünewald K., Koster A.J. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J., Lee E.Y., Lee M., Kim T., Cho Y.E. An in vivo cell-based assay for investigating the specific interaction between the SARS-CoV N-protein and its viral RNA packaging sequence. Biochem. Biophys. Res. Commun. 2019;520:499–506. doi: 10.1016/j.bbrc.2019.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Yeh S.H., Tsay Y.G., Shieh Y.H., Kao C.L., Chen Y.S., Wang S.H., Kuo T.J., Chen D.S., Chen P.J. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 2009;284:5229–5239. doi: 10.1074/jbc.M805747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Chen P.J., Yeh S.H. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe. 2014;16:462–472. doi: 10.1016/j.chom.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Liu P., Wudeck E.V., Giedroc D.P., Leibowitz J.L. SHAPE analysis of the RNA secondary structure of the Mouse Hepatitis Virus 5′ untranslated region and N-terminal nsp1 coding sequences. Virology. 2015;475:15–27. doi: 10.1016/j.virol.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., West A.M.V., Silletti S., Corbett K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020 doi: 10.1002/pro.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Gustafson C.L., Diao J., Burgner J.W., 2nd, Li Z., Zhang J., Chen J. Recombinant severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein forms a dimer through its C-terminal domain. J. Biol. Chem. 2005;280:23280–23286. doi: 10.1074/jbc.M501015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.M., Oldham M.L., Zhang J., Chen J. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 2006;281:17134–17139. doi: 10.1074/jbc.M602107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Su J.M., Samuel C.E., Ma D. Measles Virus Forms Inclusion Bodies with Properties of Liquid Organelles. J. Virol. 2019;93:e00948-19. doi: 10.1128/JVI.00948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga S., Cruz J.L., Sola I., Mateos-Gómez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuwała K., Golda A., Kabala W., Burmistrz M., Zdzalik M., Nowak P., Kedracka-Krok S., Zarebski M., Dobrucki J., Florek D. The nucleocapsid protein of human coronavirus NL63. PLoS ONE. 2015;10:e0117833. doi: 10.1371/journal.pone.0117833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Uncropped microscopy images are available from Mendeley Data: https://data.mendeley.com/datasets/88xpyp9prs/2.