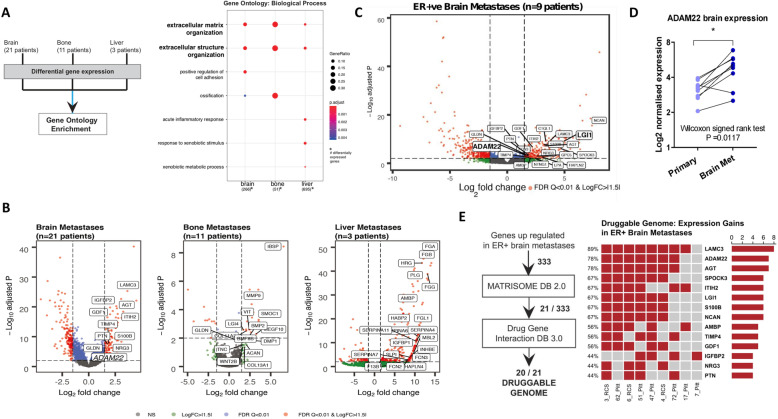
Fig. 1.
ECM is a key pathway in breast cancer metastasis and the ECM signalling protein ADAM22 promotes distant metastatic disease burden in vivo. a Sequencing reads from exome capture RNAseq of patient-matched primary breast tumour with bone metastases (n = 11 patients) and brain metastases (n = 21 patients) and from RNA seq of matched primary and metastatic liver (n = 3 patients) were mapped against human reference transcriptome GRCh38.p10. DESeq2 (exome capture RNAseq) and edgeR (RNAseq) were used to identify differential gene expression separately in primary breast tumours compared to matched brain (266), bone (51) and liver (695) metastases (log2 fold change ± 1.5 FDR < 0.05). Dotplot (right) of functional annotation of differentially expressed genes using gene ontology biological process (hypergeometric test q-value < 0.05). b Differentially expressed genes (log2 FC > 1.5, adjusted p value < 0.01) are displayed in the volcano plot. Genes were cross referenced against the Matrisome database to identify extracellular matrix (ECM) related genes (labelled in the volcano plot). c Exome-capture RNAseq of ER-positive primary breast and matched brain metastatic tissues (n = 9 patients, 18 samples). Differentially expressed genes (log2 FC > 1.5, adjusted p value < 0.01) are displayed in the volcano plot. Genes were cross referenced against the Matrisome database to identify extracellular matrix (ECM)-related genes (labelled in the volcano plot). d A significant increase in ADAM22 expression was found in brain metastases in patients in comparison to matched primary tissue, n = 9, Wilcoxon matched-pairs signed-rank test *p = 0.0117. e Oncoprint of recurrent (> 3 patients) Matrisome and Druggable Genome (Drug-Gene Interaction Database)-related gene expression gains in ER-positive brain metastases patients (n = 9) showed ADAM22 (78%) as the second-ranked druggable genome in the list