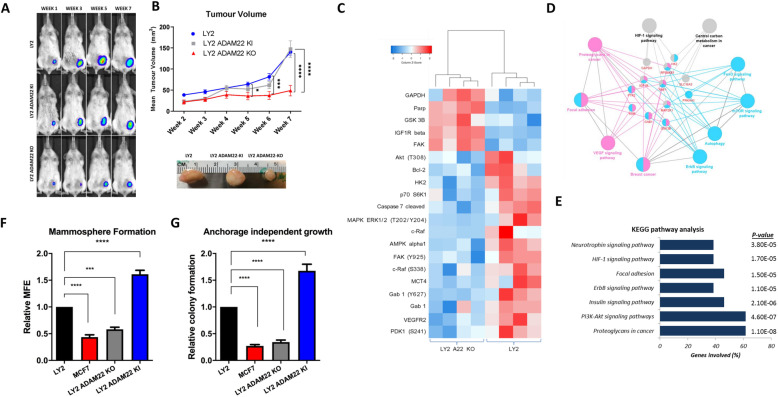
Fig. 2.
ADAM22 expression promotes metastatic potential of endocrine resistant cells via upregulation of pro-metastatic signalling pathways. a LY2 WT, LY2 ADAM22 KI and LY2 ADAM22 KO (1 × 106) cells were luciferase tagged and injected into the mammary fat pad of NOD/SCID mice (n = 8, n = 7 and n = 7, respectively) and allowed to grow to approximately 150 mm3. b Tumour growth was assessed by IVIS 15 min post luciferin injection (150 μg/g). Tumour volume was measured by weekly calliper measurements using the following formula: Volume mm3 = (length × (smallest width) × 2) × 0.5. Statistical significance was calculated using 2-way ANOVA multiple comparison test *p = 0.0395, ***p < 0.0005, ****p < 0.0001. c Heatmap from RPPA analysis showing proteins/phospho-proteins differentially expressed in LY2 and LY2 ADAM22-KO cells following 4-OHT (10− 7 M) treatment for 15 min (LIMMA adjusted p value < 0.05; red = upregulated; blue = downregulated). d Pathway analysis using ClueGo represent the common pathways from KEGG pathway analysis using the common targets from 4 or more differentially expressed proteins by ADAM22. e Top ranking KEGG pathways associated with ADAM22 dependent 4-OHT response. f ADAM22 promotes mammosphere formation. LY2, MCF7, LY2 ADAM22 KO and LY2 ADAM22 KI cells were plated in mammosphere-forming medium supplemented with 4-OHT (10− 8 M) for 5 days. Mammospheres (> 50 μm) were counted to determine the mammosphere forming efficiency (MFE). Bar graphs show relative (to LY2) MFE ± SEM from three independent experiments. Statistical significance was calculated using one-way ANOVA, ***p = 0.0001 ****p < 0.0001. g ADAM22 expression promotes anchorage independent colony formation. LY2, LY2 ADAM22 KO, MCF7 and LY2 ADAM22 KI cells cultured in an anchorage independent state for 14 days. Colonies were stained with p-iodonitrotetrazolium and counted. Bar graphs show relative (to LY2) colony formation ± SEM from three independent experiments. Statistical significance was calculated using one-way ANOVA, ****p < 0.0001