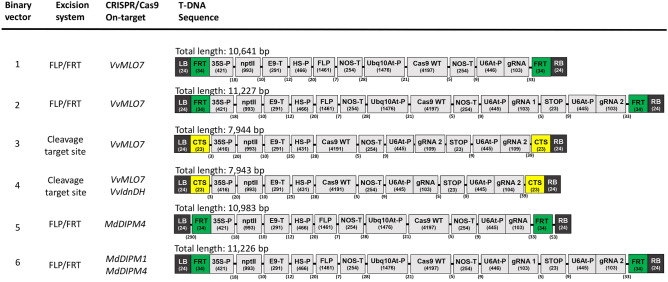
Figure 1.
Summary of binary vectors carrying self-excisable CRISPR T-DNAs used in the present study. Vectors from 1 to 4 were used for grapevine transformation while vectors 5 and 6 for apple transformation. T-DNAs contained the CRISPR/Cas9 system driven by a constitutive (Arabidopsis thaliana Ubiquitin-10 Promoter, Ubq10At-P) or an inducible (Heat Shock-Promoter, HS-P) promoter. For grapevine, the editing targets were the powdery mildew susceptibility gene VvMLO7 and the L-idonate dehydrogenase gene VvIdnDH. For apple, the editing targets were the fire blight susceptibility genes MdDIPM1 and MdDIPM4. T-DNAs were specifically designed to be self-excisable by using two different excision systems: (i) the FLP (Flippase)/FRT (Flippase Recognition Target site) recombination system and (ii) the CTS (Cleavage Target Site) recognized by the CRISPR/Cas9 system. Left and Right Borders (LB and RB); Cauliflower Mosaic Virus 35S Promoter (35S-P); Neomycin phosphotransferase II (nptII); E9 Terminator (E9-T); NopalinE Synthase Terminator (NOS-T); Crispr associated protein 9 wild-type (Cas9 WT); Arabidopsis thaliana U6 Promoter (U6At-P); guide RNA for the CRISPR/Cas9 system (gRNA); short hairpin to detach RNA-polymerase from DNA strand (STOP).