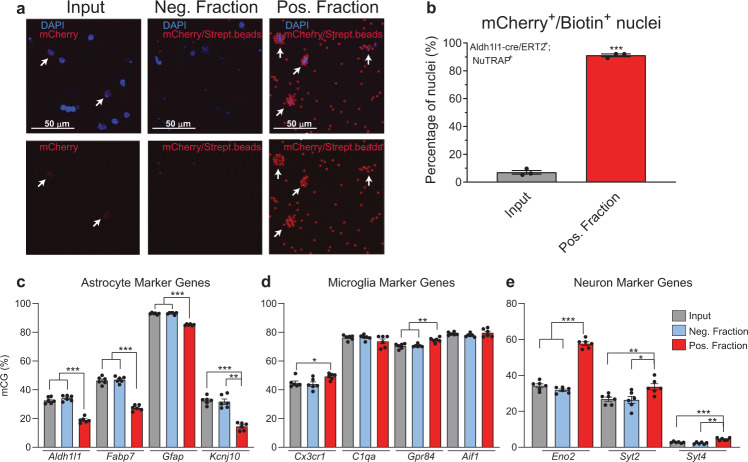
Fig. 4. Validation of astrocytic nuclei and epigenome enrichment in the Aldh1l1-NuTRAP mouse brain by INTACT-BSAS.
a Representative confocal fluorescent microscopy images from input, negative, and positive INTACT nuclei fractions. Scale bar: 50 µm. b Purity of astrocytic nuclei, expressed as average percentage ± SEM mCherry+/ Biotin+ nuclei in the positive fraction, compared to percentage ± SEM mCherry+ nuclei in the input demonstrates a high degree of specificity to the INTACT isolation (n = 3/group, ***p < 0.001 by paired t-test comparing positive fraction to input). c–e INTACT-isolated genomic DNA from Aldh1l1-NuTRAP mice was bisulfite converted and DNA methylation in specific regions of interest (promoters for neuron, astrocytes and microglia marker genes) were analyzed by Bisulfite Amplicon Sequencing (BSAS) from input, negative, and positive fractions. Hypomethylation of the astrocyte marker genes Aldh1l1, Fabp7, Gfap, and Kcnj10 in the positive fraction compared to input and negative fraction and hypermethylation of the microglial genes Cx3cr1 and Gpr84, and hypermethylation of the neuronal marker genes Eno2, Syt2, and Syt4 was observed (n = 6/group, average % mCG ±SEM, RM One-way ANOVA with Tukey’s post-hoc, *p < 0.05, **p < 0.01, ***p < 0.001).