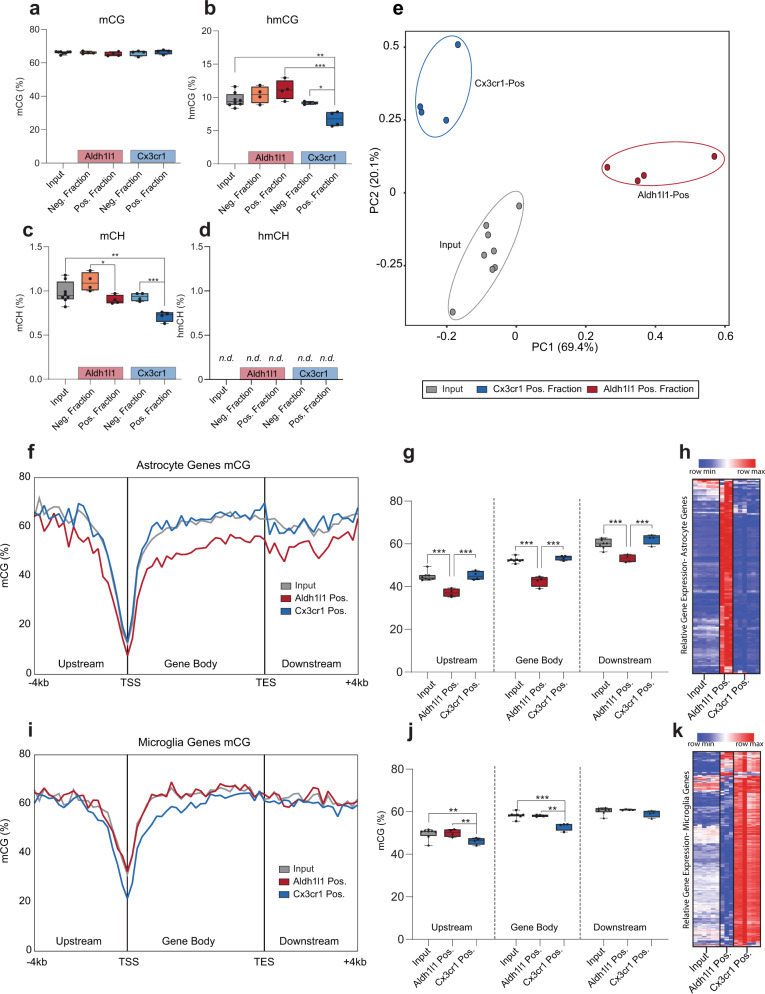
Fig. 9. DNA modification profiles of INTACT-isolated DNA from Aldh1l1-NuTRAP and Cx3cr1-NuTRAP mouse brains by WGoxBS.
INTACT-DNA samples from Aldh1l1-NuTRAP and Cx3cr1-NuTRAP brains were used for epigenome analyses. a–d Total genomic levels of mCG, hmCG, mCH, and hmCH (n = 8/input, n = 4/positive fraction; One-way ANOVA with Tukey’s multiple comparisons test, *p < 0.05, **p < 0.01, ***p < 0.001). e Principal component analysis of average gene body mCG (%) across astrocytic, microglial, neuronal, and oligodendrocytic cell marker lists separates input, Aldh1l1-NuTRAP-positive (Aldh1l1-Pos), and Cx3cr1-NuTRAP-positive (Cx3cr1-Pos) fractions. f mCG averaged over 200 nucleotide bins upstream, in the gene body, and downstream of published astrocyte genes (McKenzie)18 in the positive fraction of Aldh1l1-NuTRAP, positive fraction of Cx3cr1-NuTRAP, and input samples combined. g Average percentage mCG in the positive fraction of Aldh1l1-NuTRAP, positive fraction of Cx3cr1-NuTRAP, and input samples combined in genomic DNA upstream 4 kb of the TSS, in the gene body, and downstream 4 kb of the TES of astrocytic genes. h Hypomethylation of astrocytic gene promoters in the Aldh1l1-NuTRAP-positive fraction correlates with higher astrocytic gene expression in the Aldh1l1-positive fraction than input and Cx3cr1-NuTRAP-positive fraction. i mCG averaged over 200 nucleotide bins upstream, in the gene body, and downstream of published microglia genes18 in the positive fraction of Aldh1l1-NuTRAP, positive fraction of Cx3cr1-NuTRAP, and input. j Average percentage mCG in the positive fraction of Aldh1l1-NuTRAP, positive fraction of Cx3cr1-NuTRAP, and input DNA upstream 4 kb of the TSS, in the gene body, and downstream 4 kb of the TES of microglia genes. k Hypomethylation of microglia gene promoters in the Cx3cr1 positive fraction correlates with higher microglia gene expression. e–i n = 8/input, n = 4/positive fraction; 2-way ANOVA with Sidak’s multiple comparison test, *p < 0.05, **p < 0.01, ***p < 0.001.