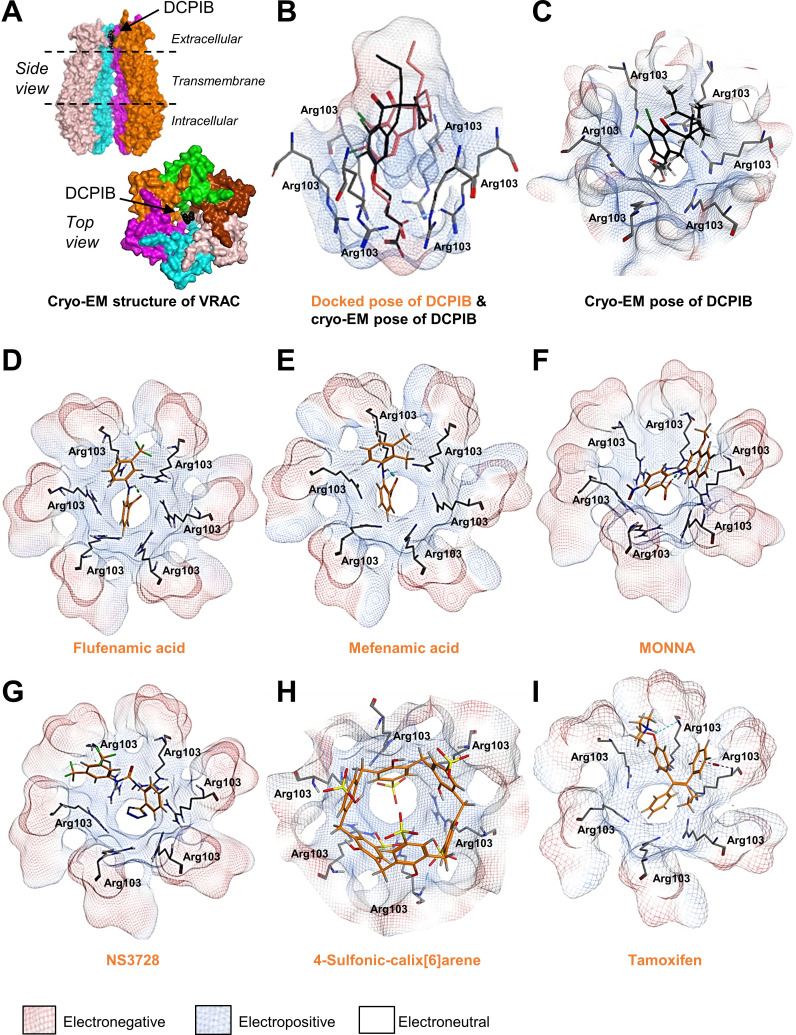
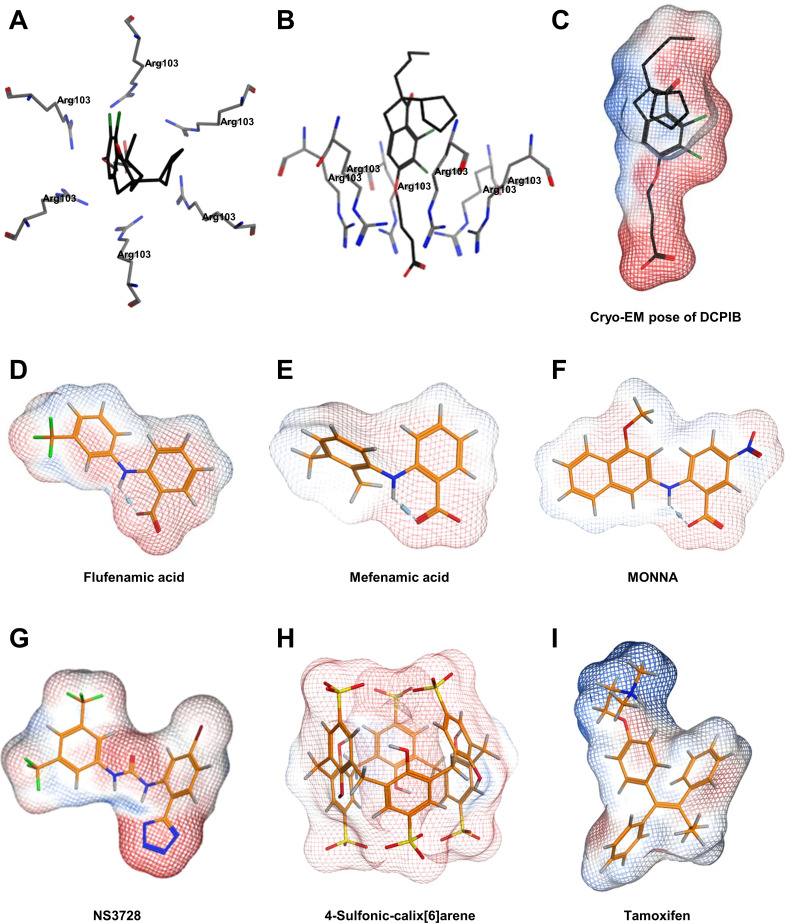
Figure 1. Modelling of proposed VRAC inhibitors on the Cryo-EM structure of LRRC8 channels.
(A) Two orthogonal views: a side view with two chains removed from the hexameric LRRC8A protein with bound DCPIB and a top view for the hexameric VRAC channel (PDB:6NZW). (B, D–I) Docked VRAC inhibitors (orange) in the VRAC Arg103 extracellular selectivity filter. Protein surface coloured according to electrostatics: electronegative (red), electropositive (blue), and electroneutral (white). (B) MM/GBVI binding energies of docked DCPIB (orange) in VRAC (−5.2 kcal mol−1) in a side view superimposed on the cryo-EM DCPIB pose (black). (C) Top-view of cryo-EM pose of DCPIB (black) in VRAC. (D) Flufenamic acid (−5.1 kcal mol−1) (E) Mefenamic acid (−5.2 kcal mol−1) (F) MONNA (−5.8 kcal mol−1) (G) NS3728 (−6.4 kcal mol−1) (H) 4-sulfonic calix[6]arene (−9.9 kcal mol−1) (I) Tamoxifen (−6.5 kcal mol−1).