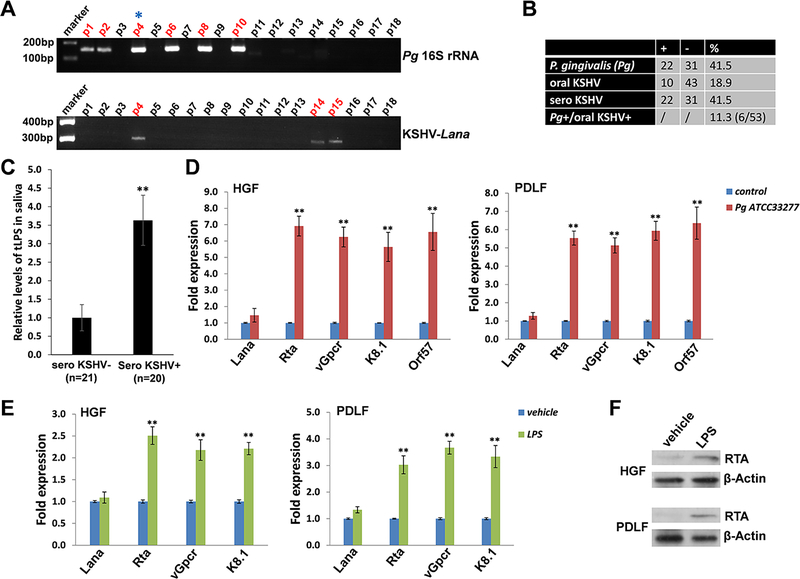
Figure 1. Clinical prevalence of P. gingivalis and KSHV shedding within patients’ saliva and induction of KSHV lytic reactivation from oral cells by P. gingivalis.
(A) Total DNA was extracted from saliva samples of cohort HIV+ patients by using QIAamp DNA Mini-kit (Qiagen), then PCR was performed using specific primers designed for 16S rRNA of P. gingivalis or KSHV-encoded major latent gene Lana, respectively. Amplicons were subsequently identified by ethidium bromide-loaded agarose gel electrophoresis and representative bands shown only. Asterisks represent double-positive patients. The KSHV seroprevalence was determined by using quantitative ELISAs as described in the Methods. (B) The results of total 53 HIV+ patients’ samples were calculated. (C) The salivary total lipopolysaccharides (LPS) levels in cohort HIV+ patients were measured by using ELISA. ** = p<0.01. (D) Oral fibroblasts were infected by purified KSHV (MOI~10) for 2 h. 24 h later, cells were treated with filtered conditioned medium from overnight P. gingivalis (Pg) ATCC33277 strain culture or fresh medium control (diluted as 1:50) for additional 48 h. qRT-PCR was used to quantify the expression of representative viral latent (Lana) and lytic genes (Rta, vGpcr, K8.1 and Orf57). (E-F) Cells were infected as (D), then incubated with Pg-derived LPS (5.0 μg/mL) for 48 h followed by qRT-PCR. Protein expression was detected using immunoblots. Error bars represent the S.D. for 3 independent experiments. ** = p<0.01. HGF: human gingival fibroblasts; PDLF: periodontal ligament fibroblasts.