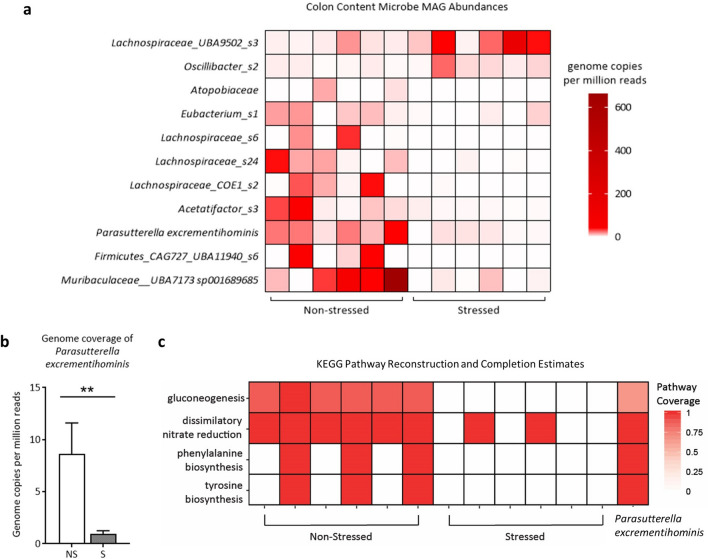
Figure 5.
GD17 colonic microbiota metagenome analyses reveal restricted bacterial abundances and metabolic pathways due to psychological stress. (a) Differentially abundant metagenome-assembled genomes (MAGs) in non-stressed and stressed samples (p < 0.05, LDA > 2.0), as identified by LEfSe, using genome copies per million reads. Taxonomy is GTDB taxonomy with the lowest possible classification listed. Some MAGs have higher level taxonomy listed for easier identification. (b) Parasutterella excrementihominis genome copies per million reads (mean ± SEM; Kruskal Wallis and Wilcoxon Signed Rank tests, p = 0.006). (c) KEGG pathway completion for select KEGG pathways in whole sample assemblies that displayed presence/absence differences between sample groups using KEGGDecoder. Samples are grouped by non-stress and stress, with the KEGG pathway completions for the consensus Parasutterella excrementihominis MAG on the far right. NS = non-stressed, S = stressed; n = 6/group; **p < 0.01.