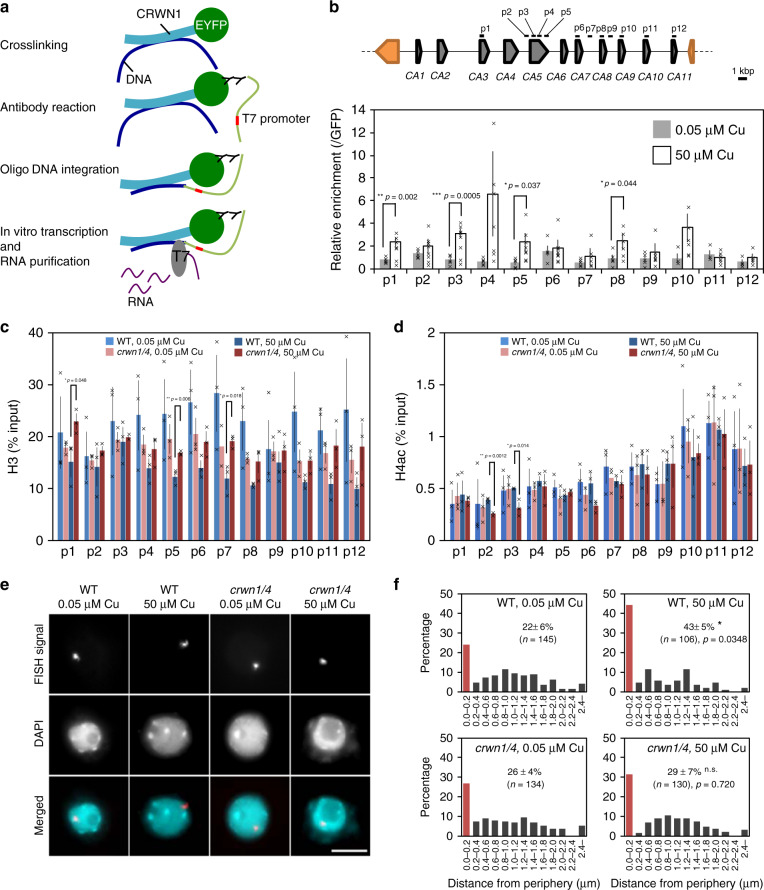
Fig. 5. CRWNs regulate the position of the CA gene locus depending on copper concentration.
a Outline of chromatin integration labeling (ChIL) method. b ChIL-qPCR analyses using 12 primer pairs described as p1 to p12. Top image indicates the position of primer pairs at the CA gene locus. Data were normalized to ChILed DNA levels in GFP control and are expressed as mean ± SEM. Significance was determined using unpaired two-sided t-test (n ≥ 4 individual experiments). Each data point represents a cross mark. c, d ChIP assay for histone H3 (c) and H4 acetylation (d) in the CA gene locus under normal and excess copper conditions. Data are expressed as mean ± SEM. Significance was determined using unpaired two-sided t-test (n = 3 individual experiments). Each data point represents a cross mark. e Visualization of the CA gene locus in the nucleus by padlock FISH. In the merged panel, FISH signals are red and DAPI signals are cyan. Scale bar = 5 µm. Data are from single representative experiments that were reproduced three times. f The distance between the CA gene locus and the nuclear edge was measured. The red bars in the histogram represent the nuclear periphery including the region 0.0 to 0.2 μm from the nuclear edge. The average percentage of the CA gene locus within the nuclear periphery with SEM from three independent replicates is shown. “n” represents the total number of FISH signals analyzed from all replicates. The CA gene locus distribution data under normal conditions were compared with those under excess copper conditions in WT and crwn1crwn4. Significance was determined using unpaired two-sided t-test.