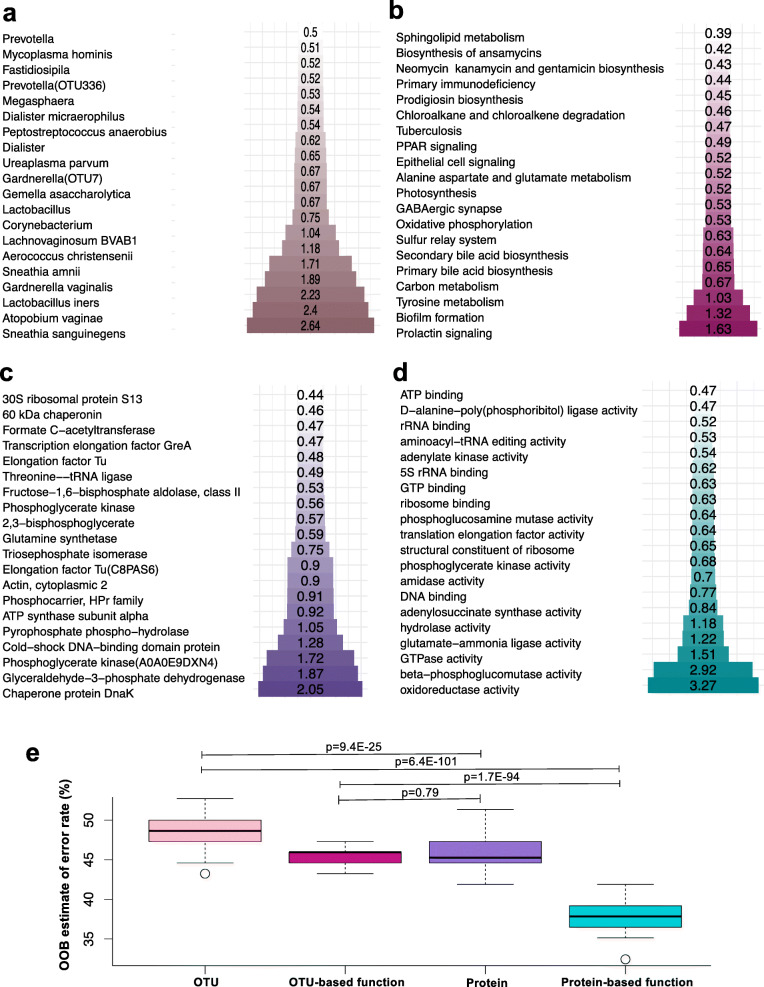
Fig. 3.
Comparison of bacterial, bacterial protein, and bacterial function relative abundance for prediction of genital inflammation. Random forest analysis was used to evaluate the accuracy of a bacterial relative abundance (determined using 16S rRNA gene sequencing; n = 74), b bacterial functional predictions based on 16 rRNA data (n = 74), c bacterial protein relative abundance (determined using metaproteomics; n = 74), and d bacterial protein molecular function relative abundance (determined by metaproteomics and aggregation of protein values assigned to the same gene ontology term; n = 74) for determining the presence of genital inflammation (low, medium, and high groups). Inflammation groups were defined based on hierarchical followed by K-means clustering of women according to the concentrations of nine pro-inflammatory cytokines [interleukin (IL)-1α, IL-1β, IL-6, IL-12p40, IL-12p70, tumor necrosis factor (TNF)-α, TNF-β, TNF-related apoptosis-inducing ligand (TRAIL), interferon (IFN)-γ]. The bars and numbers within the bars indicate the relative importance of each taxon, protein, or function based on the Mean Decrease in Gini Value. The sizes of bars in each panel differ based on the length of the labels. e Each random forest model was iterated 100 times for each of the input datasets separately, and the distribution of the out-of-bag (OOB) error rates for the 100 models was then compared using t tests. OTU operational taxonomic unit