Abstract
Background
In mammals, it is known that the estradiol-17β (E2) is mainly synthetized in ovarian granulosa cells (GCs), and the excessive apoptosis of GCs induces the follicular atresia. Many studies have implicated the essential role of KISS1, with the pro-synthetic effect of E2 and the anti-apoptotic effect on GCs, in the mammalian folliculogenesis, and several STAT4 potential binding sites were previously predicted on the promoter of KISS1 in pigs. However, the biological effects of STAT4 on GCs and the molecular regulation between STAT4 and KISS1 remained largely unknown.
Methods
Using the porcine GCs as the cellular model, the overexpression plasmid, small interfering RNA, 5′-deletion and luciferase assay were applied to investigate the molecular mechanisms for STAT4 regulating the expression of KISS1.
Results
In this study, the STAT4 negatively regulated the mRNA and protein levels of KISS1 in porcine GCs, and the mRNA level of STAT4 was observed to significantly decrease from immature to mature follicles, which was inversed with that of KISS1. The relative luciferase activity of KISS1 promoter was significantly increased with deletion of the fourth potential binding site (− 305/− 295), and ChIP further confirmed that the STAT4 bound at − 305/− 295 region of KISS1. Besides, the STAT4 significantly regulated the mRNA levels of PDK1, FOXO3 and TSC2 of PI3K signaling pathway to promote the cell apoptosis and the percentage of cells at G0/G1 phase of cell cycle in GCs. Alternatively, the STAT4 significantly decreased the mRNA levels of CYP17, 3B-HSD, 17B-33 HSD, ESR1, and ESR2, as well as the concentration of E2 in GCs. Furthermore, interfering with the expression of STAT4 was observed to significantly stimulate the pro-synthetic effect of E2 and anti-apoptotic effect of KISS1 in GCs.
Conclusions
Collectively, the STAT4 might directly target at − 305/− 295 region of KISS1 to negatively regulate the transcription of KISS1, promote the cell apoptosis via PI3K signaling pathway, suppress the synthesis of E2 through the estrogen signaling pathway in porcine GCs. These proposed works could provide useful insight in further investigations on the molecular functionalities of STAT4 and KISS1 in the folliculogenesis of mammals.
Keywords: STAT4, KISS1, Cell apoptosis, Synthesis of E2, Ovarian granulosa cells
Background
In mammals, the Kisspeptin encoded by Kiss-1 metastasis-suppressor (KISS1) gene regulates the release of gonadotropin releasing hormone at the hypothalamic level [1, 2] and is widely known for its essential role in controlling of the initiating of puberty [3, 4]. Recently, the Kisspeptin has been reported to show the local roles in the ovaries of humans [5], pigs [6, 7], rats [8], hamsters [9], dogs [10], and cats [11]. It is observed that the KISS1−/− mice fail to undergo pubertal transition and show the absence of mature follicles [12, 13]. In the ovaries of rats [8] and dogs [10], the mRNA of KISS1 expresses in a distinctive stage-specific pattern throughout the estrous cycle. In cats, the mRNA level of KISS1 in follicular stage expresses higher than in luteal stage [11]. In pigs, the mRNA level of KISS1 is found to significantly increase from immature to mature follicles [6]. These observations highly implicate the essential role of KISS1 in the mammalian folliculogenesis.
Previous studies have suggested that the signal transducer and activator of transcription (STAT) family of cytosolic transcription factors [14, 15] appears to be phosphorylated to regulate the apoptosis and differentiation of various cells [16]. Many studies have reported that the STAT proteins highly express in granulosa cells (GCs) [17, 18] and play an essential role in folliculogenesis [19] and female reproduction [17, 20]. The knockout of GC-specific STAT3 significantly reduces the litter sizes of mice [20]. The levels of phosphorylated STAT3 in GCs of subordinate follicles are markedly higher than dominant follicles in bovines [21, 22]. More recently, the STAT4 has been linked to the ovarian endometriosis in women [23]. These results indicate that STATs may play an important role in mammalian folliculogenesis.
In mammals, it is known that estradiol-17β (E2) is predominantly synthetized in GCs, and the apoptosis of GCs plays an important role in deciding the fate of follicles [24–26]. The abnormal apoptosis of GCs induces follicular atresia [27] and exhibits a negative influence on oocyte quality [28] and fertility [29, 30]. The phosphatidylinositol 3-OH-kinase (PI3K) signaling pathway has been reported to promote cell survival and suppress apoptosis in mammalian GCs [31, 32] and is crucial for follicle growth [33]. Previously, we have found that the KISS1 suppresses the apoptosis and cycle entry through PI3K signaling pathway in GCs to promote the maturation of follicles [6]. Besides, the mice deficient in STAT4 demonstrate that STAT4 induces the expression of genes involved in the proliferation and apoptosis of diverse cells [34, 35]. Moreover, several STAT4 putative binding locations were predicted on the promoter of KISS1 in pigs. Therefore, we hypothesized that STAT4 might bind at the promoter of KISS1, regulate the cell apoptosis and E2 secretion in GCs, and then promote the follicular development.
In this study, the molecular mechanism for the regulation between STAT4 and KISS1 was first determined, and then their biological functions were explored on PI3K signaling pathway, cell cycle entry, cell apoptosis, estrogen signaling pathway, and E2 secretion in porcine GCs. These works will provide useful information on the molecular mechanism of STAT4-mediated-KISS1 in mammalian folliculogenesis.
Materials and methods
Ethics approval
All experiments in the present study were performed in accordance with the guidelines of the Animal Care and Use Committee of South China Agricultural University Guangzhou, China (Approval Number: SCAU#2013–10).
Animals and sample preparation
The details of animals and sample preparations were previously described in our study [6]. Briefly, three Landrace × Yorkshire crossbred gilts with the exhibitions of the first estrus were selected [36, 37]. At least three of follicles within 8–10 mm and 5–7 mm were respectively collected and were respectively considered as the mature and immature follicles. These pigs were reared in the same conditions and fed the same diet daily. The collected follicles were frozen in liquid nitrogen and then transferred to store at − 80 °C fridge for further using.
Prediction of potential STAT4 binding sites at the promoter of KISS1
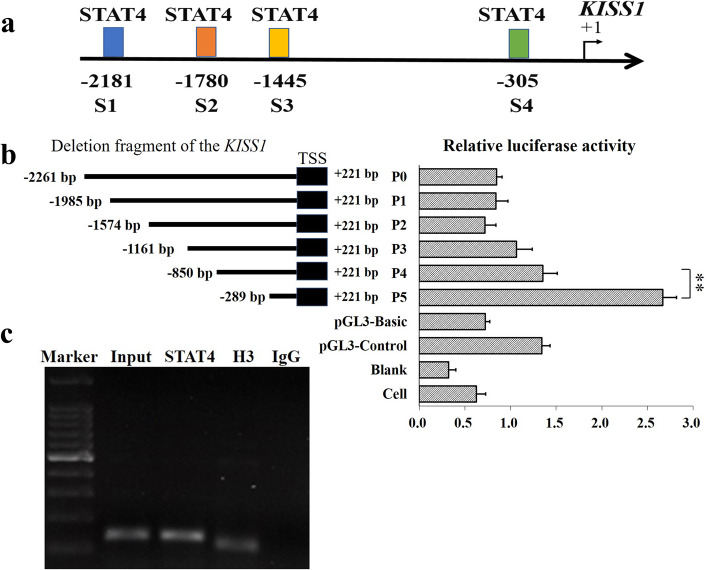
The promoter sequences of porcine KISS1 (upstream 2.5 kb) were download and acquired from NCBI (https://www.ncbi.nlm.nih.gov/gene/100145896). TFBIND [38], Biobase (http://gene-regulation.com/pub/programs/alibaba2/index.html), Jaspar [39] and Research (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) were applied to predict the putative and potential binding site of STAT4. The putative binding sites of STAT4 predicted by all of those four tools were selected for further using. The locations of these potential binding sites on the promoter of KISS are shown in Fig. 2a.
Fig. 2.
STAT4 binds at − 305/− 295 region of KISS1. a Predictions of the potential binding sites of STAT4 at the promoter of KISS1 in pigs; b Relative luciferase activity of KISS1 promoter after 5′ deletion of the potential binding sites of STAT4; c Confirmation of STAT4 binding at − 305/− 295 by ChIP. ** indicates P < 0.01. Data were represented as means ± SD
Construction of KISS1 5′ deletion and luciferase assay
The total genomic DNA of porcine ovaries was extracted and used as the template. PCR was conducted by using PrimerSTAR® (TaKaRa, Dalian, Liaoning, China) enzyme to obtain the promoter sequences of KISS1 (2482 bp). Then PCR products were purified to combine with pMD-18 T and transformed into competent cells DH5α. These DH5α were inoculated on ampicillin-containing lysogeny broth plates at 37 °C for overnight, and then the monoclonal bacteria was selected and amplified overnight at 37 °C shaker. The plasmids of bacteria were collected and extracted. The correct plasmid after sequencing identification was named T-KISS1. Then T-KISS1 was used as a template to design other five deletion fragments. The same method was used to acquire plasmids of each deletion fragment including P0 (− 2261/+ 221), P1 (− 1985/+ 221), P2 (− 1574/+ 221), P3 (− 1161/+ 221), P4 (− 850/+ 221), and P5 (− 289/+ 221) with SacI and SmaI cleavage sites (Table 1). Finally, each deletion fragment was cloned into pGL3-Basic. According to Promega’s dual luciferase reporter assay kit (Promega, Madison, WI, USA) and previous study [40], the BioTek Synergy 2 multifunctional microplate reader (BioTek, Winooski, VT, USA) was used for fluorescence detection. The relative expression of firefly luciferase to renilla luciferase was targeted as the fragment activity. The primers used in this study are presented in Table 1.
Table 1.
Primers for Construction of KISS1 5′ Deletion
| Name | Sequence | Product (bp) | Accession number |
|---|---|---|---|
| P0(−2261/+ 221) | F: CGCGAGCTCCCTCACTCACCAGCCTGTTTC | 2482 | NM_001134964.1 |
| R: CCCCCGGGGGCCAGCCAGTCTTAGGTTTCCATTA | |||
| P1(−1985/+ 221) | F: CGCGAGCTCGACATCCCTCACTCCCTACTACCC | 2206 | NM_001134964.1 |
| ditto | |||
| P2(−1574/+ 221) | F: CGCGAGCTCTGGGTTTCAGGGTATCACAGAGC | 1795 | NM_001134964.1 |
| ditto | |||
| P3(−1161/+ 221) | F: CGCGAGCTCGGCTCGCCAGTGGTTTATCTTT | 1382 | NM_001134964.1 |
| ditto | |||
| P4(−850/+ 221) | F: CGCGAGCTCTAAGGGTTATGAGAGCAAGCAGGAT | 1071 | NM_001134964.1 |
| ditto | |||
| P5(−289/+ 221) | F: CGCGAGCTCTGCTATTCAGACTCATCCCTCCACT | 510 | NM_001134964.1 |
| ditto |
* The underlined is enzyme-cutting sites
Culture of porcine GCs in vitro
The porcine ovarian GCs were cultured as previously described [40, 41]. Briefly, the ovaries, which were collected from a local slaughterhouse (Guangzhou) for pigs, were transferred to laboratory in PBS (Invitrogen, Shanghai, China). Subsequently, the GCs were collected and isolated from 5 to 7 mm follicles, and then these GCs were washed twice with PBS. The GCs were cultured and seeded into 25-cm2 flasks within 37 °C and 5% CO2 in DMEM (Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (Hyclone), 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Real - time quantitative PCR analysis
The pcDNA3.1-Basic (200 ng), pcDNA3.1-STAT4 (200 ng), pcDNA3.1-KISS1 (200 ng), siRNA-STAT4 (50 nM), or siRNA-NC (50 nM) was transfected into the cells for 48 h respectively, while GCs covered 30–50% of one well. The total RNA was collected and extracted for at least triplicates per group by using TRIzol reagent (TaKaRa, Tokyo, Japan) and then reverse-transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA) for mRNAs. The Maxima SYBR Green qRT-PCR Master Mix (2×) (Thermo Scientific) was used to quantify the relative expression levels of mRNAs in a LightCycler Real-Time PCR system. The relative mRNA expression levels of genes were calculated using the 2-ΔΔct strategy with GAPDH as the endogenous controls. The primer sequences are listed in Table 2.
Table 2.
Primers of RT-PCR, ChIP and coding sequences cloning
| Name | Sequence | Product (bp) | Accession number |
|---|---|---|---|
| CDS-STAT4 | F: GGGGTACCATGTCTCAGTGGAATCAAGTC | 2266 | NM_001197305.1 |
| R: CCAAGCTTTCAGTCTGAGTCAGGTCCTT | |||
| CDS-KISS1 | F: CCGAATTCATGAATGCACTGGTTTTTTGG | 431 | NM_001134964.1 |
| R: GGCGCCGGCGAGTCAGAGCGGGCCGCGGAA | |||
| qRT-PCR-KISS1 | F: AACCAGCATCTTCTCACCAGG | 192 | NM_001134964.1 |
| R: CTTTCTCTCCGCACAACGC | |||
| qRT-PCR-STAT4 | F: TTGTCTGCTCTACCATTCGCTG | 182 | NM_001197305.1 |
| R: TAACCTTTGTCTCCCCTTTCTG | |||
| qRT-PCR-GAPDH | F: TCCCGCCAACATCAAAT | 163 | XM_021091114.1 |
| R: CACGCCCATCACAAACAT | |||
| qRT-PCR-PIK3CG | F: AACGGGCTTTGAGATAGTGAA | 184 | NM_213939.1 |
| R: AAGTTGCTTGGTTGGTGGATA | |||
| qRT-PCR-PIK3C1 | F: CAAGTGAGAATGGTCCGAATG | 152 | NM_006218.3 |
| R: GTGGAAGAGTTTGCCTGTTTT | |||
| qRT-PCR-PDK1 | F: AAATCACCAGGACAGCCAATA | 190 | NM_001159608.1 |
| R: CTTCTCGGTCACTCATCTTCAC | |||
| qRT-PCR-FOXO3 | F: ACAAACGGCTCACTCTGTCCCA | 85 | NM_001135959.1 |
| R: GAACTGTTGCTGTCGCCCTTATC | |||
| qRT-PCR-TSC2 | F: CGAGGTGGTGTCCTACGAGAT | 115 | XM_005655162.3 |
| R: GAGCAGGCGTTCAATGATGTT | |||
| qRT-PCR-BAD | F: AGTCGCCACTGCTCTTACCC | 172 | XM_021082883.1 |
| R: TCTTGAAGGAACCCTGGAAATC | |||
| qRT-PCR-Star | F: GGAAAAGACACAGTCATCACCCAT | 121 | NM_213755.2 |
| R: CAGCAAGCACACACACGGAAC | |||
| qRT-PCR-CYP17 | F: AAGCCAAGACGAACGCAGAAAG | 228 | NM_214428.1 |
| R: TAGATGGGGCACGATTGAAACC | |||
| qRT-PCR-3B-HSD | F: GGGGCTTCTGTCTTGATTCCA | 284 | NM_001004049.2 |
| R: GGTTTTCAGTGCTTCCTTGTGC | |||
| qRT-PCR-17B-HSD | F: CCCAACGCAGGAGACTCAAAAT | 149 | NM_214306.1 |
| R: CCAGAGCCCATAACGAAGACAGA | |||
| qRT-PCR-CYP19A | F: GCTGGACACCTCTAACAACCTCTT | 91 | NM_214430.1 |
| R: TTGCCATTCATCAAAATAACCCT | |||
| qRT-PCR-ESR1 | F: GATGCCTTGGTCTGGGTGAT | 124 | XM_003468423.2 |
| R: AGTGTTCCGTGCCCTTGTTA | |||
| qRT-PCR-ESR2 | F: AAGGGAAAAGGAGGATGGGACA | 202 | NM_0010011533.1 |
| R: CAGATAGGGACTGCGTGGAGGT | |||
| ChIP-STAT4 | F: CCTTGCCCACTTCACTCCAC | 153 | NM_001197305.1 |
| R: AGGACAGAAGGAATCGAGGGA | |||
| ChIP-GAPDH | F: GATGTCCTGAGCCCCTACAG | 102 | XM_021091114.1 |
| R: GGTAGGTGATGGGGACTGAG |
* The underlined is enzyme-cutting sites
Cell apoptosis assay
The coding sequences of STAT4 (Gene ID: 397261, accession number: NM_001197305.1) and KISS1 (Gene ID: 100145896, accession Number: NM_001134964.1) were cloned into pcDNA3.1(+) (ThermoFisher, Guangzhou, China) with the restrictive enzymes of KpnI and xhoI for STAT4; EcoRI and NotI for KISS1. The sequences of the primers for these coding sequences were shown in Table 2. STAT4-siRNA-1, STAT4-siRNA-2, STAT4-siRNA-3 and Scrambled-siRNA were synthesized and purified by RiboBio Co. Ltd. (Guangzhou, China).
According to our previous studies [40, 41], the cell apoptosis was detected to by using an Annexin V-FITC Apoptosis Detection Kit (BioVision, Milpitas, CA, USA). Briefly, when GCs covered 30–50% of the triplicate in 6-well plates at 24 h prior to transfection, pcDNA3.1-STAT4, pcDNA3.1-KISS1, pcDNA3.1-Basic, STAT4-siRNA, and Scrambled-siRNA were transfected into the cells for 48 h, respectively. Then the transfected cells were harvested and treated by using Annexin V-FITC mix and were analyzed in a flow cytometer (Becton Dickinson Co., San Jose, CA, USA). All experiments were performed at least triplicate.
Cell cycle analysis
The analysis of cell cycle was according to our previous studies [40, 41]. When GCs covered 30–50% of one well, pcDNA3.1-STAT4, pcDNA3.1-Basic, STAT4-siRNA, or Scrambled-siRNA was transfected into GCs for 48 h, respectively. The transfected cells were collected and washed twice with ice-cold PBS. These cells were resuspended using a propidium iodide/RNase A solution at 37 °C for 30 min in the dark. Then these cells were analyzed by flow cytometry (Becton Dickinson Co., San Jose, CA, USA).
ELISA for measurements of steroid hormones
After transduction with pcDNA3.1-STAT4, pcDNA3.1-KISS1, pcDNA3.1-Basic, STAT4-siRNA, and Scrambled-siRNA for 48 h, the concentrations of E2 in the culture supernatants were measured with ELISA kits (Beijing north institute of biological technology, Beijing, China) according to our previous studies [40, 41].
Data analysis
Data were expressed as means ± standard deviation (SD) of repeated experiments. The student’s t-test (two-tailed) was used to detect the significance of mean differences between two groups by using R software (version-3.4.3, https://www.r-project.org/) in this study. * indicates P < 0.05; ** indicates P < 0.01.
Results
STAT4 inhibits the mRNA and protein expressions of KISS1 in porcine GCs
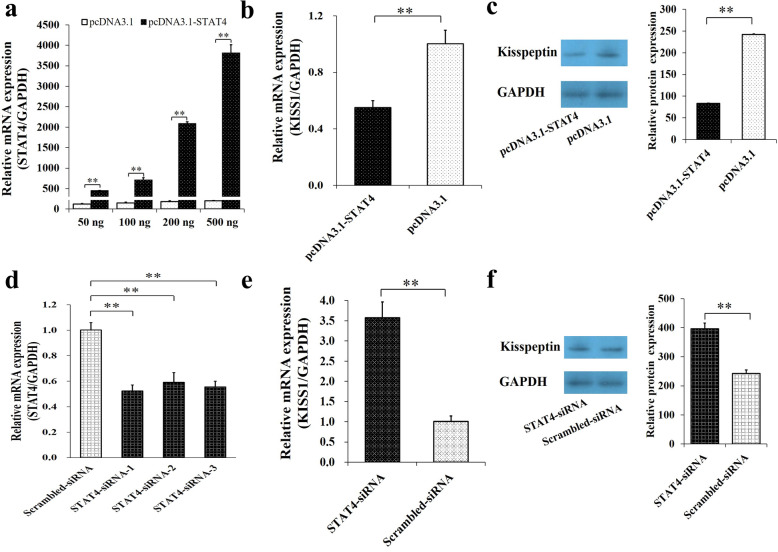
The mRNA level of STAT4 was found to be increasing with the concentration of overexpression plasmid (pcDNA3.1-STAT4) for STAT4 (Fig. 1a), and 200 ng plasmid of pcDNA3.1-STAT4 was selected for further using on the considerations for cellular tolerance. The overexpression of STAT4 significantly downregulated the mRNA (Fig. 1b, P < 0.01) and protein levels (Fig. 1c, P < 0.01) of KISS1, compared to the control group. Besides, three STAT4-specific small interfering RNA (siRNA) and a negative control (Scrambled-siRNA) were transfected into GCs to evaluate the knockdown efficiency for STAT4 (Fig. 1d), and STAT4-siRNA2 was observed to show the highest performance and then was selected to inhibit the expression of STAT4 in GCs (Fig. 1d). We found that interfering with the expression of STAT4 significantly increased the mRNA (Fig. 1e, P < 0.01) and protein levels (Fig. 1f, P < 0.01) of KISS1, compared to the control group. These results indicated that STAT4 negatively regulated the mRNA and protein expressions of KISS1 in porcine GCs.
Fig. 1.
STAT4 inhibits the mRNA and protein levels of KISS1. a Relative expression of STAT4 against the different concentrations of pcDNA3.1-STAT4 plasmid; The mRNA (b) and protein (c) expressions of KISS1 were depressed by pcDNA3.1-STAT4; d Relative mRNA expression of STAT4 knockdown by three siRNAs; The mRNA (e) and protein (f) expressions of STAT4 were stimulated by STAT4-siRNA. ** indicates P < 0.01. Data were represented as means ± SD. siRNA: small interfering RNA; Scrambled-siRNA: a siRNA negative control
STAT4 binds at − 305/− 295 region of KISS1
Four potential binding sites of STAT4 were found at the promoter of KISS1 (Fig. 2a), indicating that the STAT4 might directly target at KISS1 to regulate the expression of KISS1. To study the molecular regulation and mechanism between STAT4 and KISS1, the 5′-deletion reporter was built and constructed for KISS1 (Fig. 2b). The deletion of S1 (− 2181/− 2171), S2 (− 1780/− 1770), and S3 (− 1445/− 1435) did not show significant changes on the relative luciferase activity of P1, P2, and P3, compared to P0 (Fig. 2b). However, compared to P0, P1, P2, P3, and P4, the deletion of the fourth potential binding site (− 305/− 295) significantly increased the relative luciferase activity (P5) (Fig. 2b). Moreover, ChIP further identified that STAT4 bound at − 305/− 295 in porcine GCs (Fig. 2c). These results demonstrated that STAT4 directly bound at − 305/− 295 region of KISS1 to negatively regulate the transcription of KISS1 in porcine GCs.
STAT4 promotes the apoptosis of porcine GCs
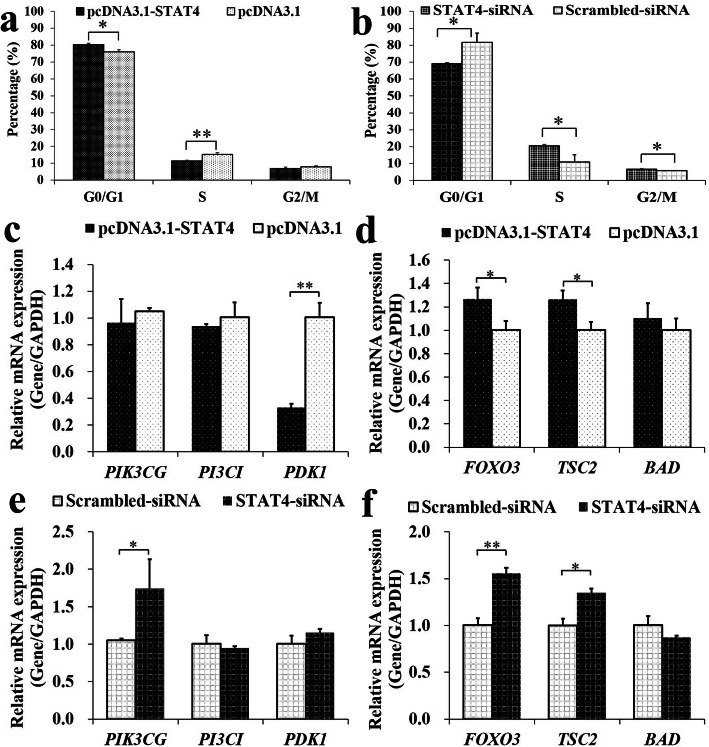
To further identify the biological function of STAT4 on cell cycle and cell apoptosis, pcDNA3.1-STAT4 or STAT4-siRNA was transfected into porcine GCs. After the analysis of cell cycle, the pcDNA3.1-STAT4 was observed to significantly upregulate the percentage of cells in the stage of G0/G1 (Fig. 3a) and downregulate the percentage of cells in the stage of S, but the STAT4-siRNA was found to significantly downregulate the percentage of cells in the stage G0/G1 and upregulate the percentage of cells in the stage of S (Fig. 3b).
Fig. 3.
Biological Effects of STAT4 on cell cycle and PI3K signaling pathway in GCs. a Effects of pcDNA3.1-STAT4 on the cell cycle stage; b Effects of STAT4-siRNA on the cell cycle stage; c Relative mRNA expressions of PIK3CG, PI3C1, and PDK1 after the treatment by pcDNA3.1-KISS1; d Relative mRNA expressions of FOXO3, TSC2, and BAD after the treatment by pcDNA3.1-KISS1; e Effects of KISS1-siRNA on the relative mRNA levels of PIK3CG, PI3C1, and PDK1; f Effects of KISS1-siRNA on the relative mRNA levels of FOXO3, TSC2, and BAD. ** indicates P < 0.01; * indicates P < 0.05. Data were represented as means ± SD
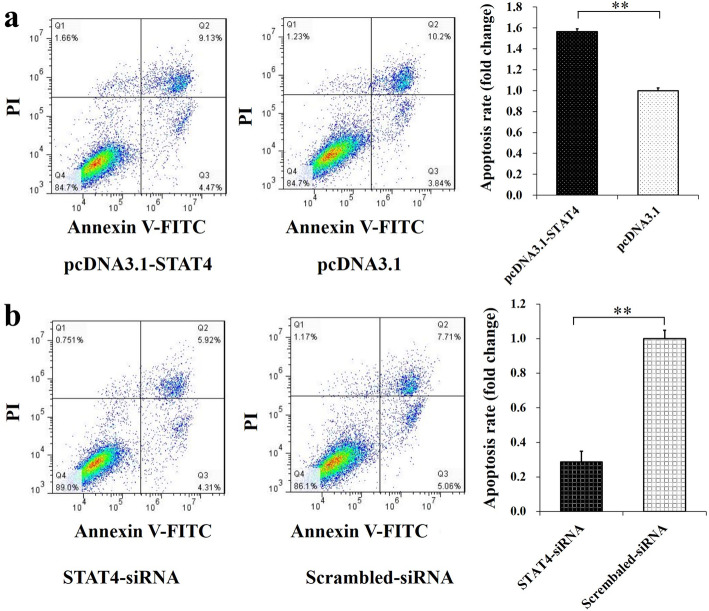
Furthermore, the pcDNA3.1-STAT4 was found to downregulate the mRNA of PDK1 (Fig. 3c) and increased the mRNA levels of FOXO3 and TSC2 (Fig. 3d) but did not show significant effects on the mRNA levels of PIK3CG, PI3C1, and BAD (Fig. 3c,d). Besides, the STAT4-siRNA markedly upregulated the mRNA levels of PIK3CG, FOXO3, and TSC2 (Fig. 3e,f), but did not show significant effects on the mRNA levels of PI3C1, PDK1, and BAD (Fig. 3e,f). Moreover, the pcDNA3.1-STAT4 was observed to significantly promote the cellular apoptosis (Fig. 4a), and the STAT4-siRNA significantly inhibited the apoptosis in porcine GCs (Fig. 4b). These results suggested that STAT4 might disturb the cell cycle and promote cell apoptosis through PI3K signaling pathway in porcine GCs.
Fig. 4.
STAT4 promotes the apoptosis of GCs. The pcDNA3.1-STAT4 decreased cell apoptosis rate (a), but the STAT4-siRNA increased cell apoptosis rate of GCs (b); The cell apoptosis was assessed by Annexin V-FITV/PI. The cells in lower right quadrant were annexin-positive/PI-negative early apoptotic cells. The cells in the upper right quadrant were annexin-positive/PI-positive late apoptotic cells. Compared to the blank group, the fold change of the percentage of cells undergoing early and late apoptosis were presented in the barplot. ** indicates P < 0.01. Data were represented as means ± SD
STAT4 inhibits the synthesis of E2 in porcine GCs
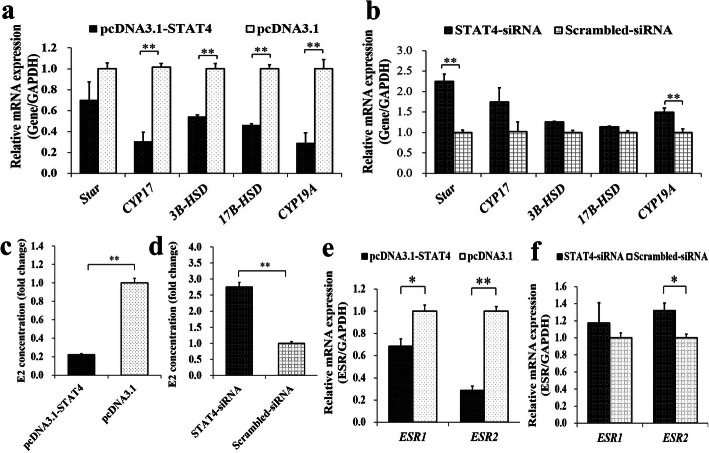
To investigate the biological effect of STAT4 on the synthesis of E2 in porcine GCs, the pcDNA3.1-STAT4 or STAT4-siRNA was transfected into porcine GCs (Fig. 5). We found that the overexpression of STAT4 significantly increased the mRNA levels of CYP17, 3B-HSD, 17B-HSD and CYP19A (Fig. 5a), and STAT4-siRNA significantly downregulated the mRNA levels of Star and CYP19A (Fig. 5b). Furthermore, the pcDNA3.1-STAT4 (Fig. 5c) or STAT4-siRNA (Fig. 5d) significantly decreased or increased the concentration of E2 in porcine GCs, respectively. Additionally, the pcDNA3.1-STAT4 significantly down-regulated the mRNA levels of ESR1 and ESR2 (Fig. 5e), and the STAT4-siRNA appeared to increase the mRNA expressions of ESR1 and ESR2 (Fig. 5f).
Fig. 5.
STAT4 inhibits the synthesis of E2 in porcine GCs. Effects of pcDNA3.1-STAT4 (a) and STAT4-siRNA (b) on the relative mRNA levels of Star, CYP17, 3B-HSD, 17B-HSD, and CYP19A; c Concentrations of E2 was depressed by pcDNA3.1-STAT4; d Concentrations of E2 was stimulated by STAT4-siRNA; e Effects of pcDNA3.1-STAT4 on the relative mRNA levels of ESR1 and ESR2; f Effects of STAT4-siRNA on the relative mRNA levels of ESR1 and ESR2. ** indicates P < 0.01; * indicates P < 0.05. Data were represented as means ± SD
STAT4 suppresses the effects of KISS1 on cell apoptosis and synthesis of E2 in GCs
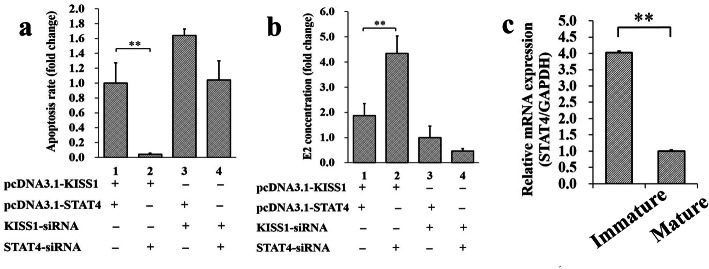
To further characterize the biological effect of STAT4 on the cellular functions of KISS1, the pcDNA3.1-STAT4, STAT4-siRNA, pcDNA3.1-KISS1, and KISS1-siRNA were transfected into porcine GCs (Fig. 6). We found that the fold change of group 1 (pcDNA3.1-STAT4 + pcDNA3.1-KISS1) was significantly higher than group 2 (STAT4-siRNA + pcDNA3.1-KISS1) (Fig. 6a) for the cell apoptosis rate, and the synthetic capacity of E2 of group 1 (pcDNA3.1-STAT4 + pcDNA3.1-KISS1) was significantly lower than group 2 (STAT4-siRNA + pcDNA3.1-KISS1) (Fig. 6b). These results indicated that STAT4 might repress the anti-apoptotic effect and the synthetic capacity of E2 of KISS1 in ovarian GCs. Moreover, the mRNA level of STAT4 was observed to significantly decrease from immature to mature follicles (Fig. 6c), which was inversed with that of KISS1 during the follicular maturation [6].
Fig. 6.
STAT4 suppresses the effects of KISS1 on cell apoptosis and synthesis of E2 in GCs. a STAT4 repressed the anti-apoptotic effect of KISS1 in ovarian GCs; b STAT4 suppressed the synthetic capacity of E2 of KISS1 in ovarian GCs; c Changes of STAT4 mRNA expression during follicular maturation. ** indicates P < 0.01; * indicates P < 0.05. Data were represented as means ± SD
Discussion
A series of studies have recently recommended that the KISS1 might get involved in folliculogensis and controlling the maturation of follicles in mammals [8, 11, 42]. In humans, the level of Kisspeptin is higher in polycystic ovarian syndrome patients than control group, suggesting that the Kisspeptin may be a potential biomarker for the diagnosis of polycystic ovarian syndrome [43]. It is widely known that E2 is mainly synthetized and produced in GCs [44, 45], and the survival and proliferation of GCs directly stimulate the synthesis of E2 [46, 47] and facilitate the follicular maturation [48]. The abnormal apoptosis of GCs appears to provoke the emergence of ovarian endometriosis [49, 50]. Compared with other ovarian cells, the mRNA level of KISS1 significantly expressed higher in the GCs [11]. The lower expression of KISS1 in GCs of humans may cause the abnormal follicle development [5]. Moreover, we previously find that KISS1 stimulates the synthesis of E2 and suppresses cell cycle entry as well as cell apoptosis in porcine GCs [6], while the expression level of KISS1 mRNA significantly increased from immature to mature follicles [6]. These results indicate that the KISS1 might regulate the apoptosis of GCs and the synthesis of E2 to promote the follicular development in mammals.
Although the mRNA level of STAT4 increased along with the concentration of pcDNA3.1-STAT4 at 50 ng, 100 ng, 200 ng and 500 ng, as shown in Fig. 1a, 200 ng of pcDNA3.1-STAT4 was finally selected for further using on the considerations for cellular tolerance, and 200 ng of pcDNA3.1-STAT4 was at the similar concentration level with 50 nM of siRNAs, which was suggested by the providers and instructions. In this study, there were four putative binding sites of STAT4 at the promoter of KISS1 (Fig. 2a). We found that the deletion of the fourth putative binding site (− 305/− 295) dramatically increased the relative luciferase activity of P5, compared to P0, P1, P2, P3, and P4 (Fig. 2b), and the ChIP was further applied to confirm that STAT4 bound at − 305/− 295 of KISS1 (Fig. 2c). Besides, the overexpression of STAT4 significantly decreased the mRNA (Fig. 1b) and protein levels (Fig. 1c) of KISS1, and interfering with the expression of STAT4 significantly increased the mRNA (Fig. 1e) and protein levels (Fig. 1f) of KISS1 in porcine GCs. Moreover, the mRNA levels of STAT4 (Fig. 6c) were observed to inverse with that of KISS1 [6] from immature to mature follicles. These observations suggested that the STAT4 might directly target at − 305/− 295 of KISS1 to negatively control the transcription of KISS1 in porcine ovarian GCs.
Accumulating evidence implicates the critical role of PI3K signaling pathway in the survival and apoptosis of ovarian GCs in mammals [32, 33], and the PI3K signaling pathway is suggested to be highly associated with the progression from endometriosis to ovarian cancer [51]. Previous studies recommend that PDK1-deficient in oocytes causes the depletions of the majority of primordial follicles around the first estrus [52]. The DNA structure variation of PIK3CG may strike the occurrence of ovarian cancer [53]. In early adulthood of mice, the TSC2-deficient in oocytes depletes follicles and results in premature ovarian failure [54]. In mice, the FOXO3-deficient in GCs inhibits the ovarian follicular growth [55]. The PIK3C1 gene, which encodes an isoform of the catalytic subunit of PI3K, is suggested to regulate the follicle survival [32] and involve in the cellular proliferation and apoptosis of ovarian cancer in mammals [56]. The BAD gene, known as BCL2 associated agonist of cell death, is found to regulate the apoptosis of ovarian GCs to reduce progesterone levels in sheep [57]. In this study, herein, the effects of STAT4 on the expressions of PIK3CG, PIK3C1, PDK1, FOXO3, TSC2 and BAD from PI3K signaling pathway were determined (Fig. 3c-f). We found that the pcDNA3.1-STAT4 down-regulated the mRNA of PDK1 (Fig. 3c) and up-regulated the mRNA level of FOXO3 and TSC2 (Fig. 3d), and the STAT4-siRNA significantly increase the mRNA expressions of PIK3CG, FOXO3, and TSC2 (Fig. 3e,f). Additionally, the analysis of Annexin V-FITC flow cytometry showed that the pcDNA3.1-STAT4 promoted the apoptosis of GCs (Fig. 4a), and the STAT4-siRNA inhibited the apoptosis of GCs (Fig. 4b). Moreover, the pcDNA3.1-STAT4 and STAT4-siRNA were found to increase and decrease the number of GCs at phase of G0/G1 in the cell cycle, respectively (Fig. 3a,b). These results are in according to the study that shSTAT4 lentivirus transduces more cells in G0/G1 phase and inhibits the proliferation of vascular smooth muscle [58]. These findings indicated that the STAT4 involved in PI3K signaling pathway to suppress cell cycle entry and promote cell apoptosis in porcine GCs.
The expression of Star gene, which promotes the conversion of cholesterol into pregnenolone, is enhanced by androgen in rat GCs [59], and the KISS1 is likely to trigger the expression of Star in the granulosa lutein cells of humans [60]. The CYP17 gene encodes one member of the cytochrome P450 enzymes that produce androgens and estrogens, as well as impacting follicular development [61]. The 3B-HSD gene encodes the enzyme that catalyzes the oxidative conversion and regulates the production of progesterone and E2 in GCs of cattle [62]. In women, the variants in CYP19A gene yielded fewer mature follicles [63]. The 17B-HSD converts the estrone to more active estrogens, and the disorders of 17B-HSD, estrogen receptors and estrogens cause the prevalence of the ovarian endometriosis [64]. Therefore, in this study, the impacts of STAT4 on several genes regulated the synthesis of E2 were investigated (Fig. 5), including Star, CYP17, 3B-HSD, 17B-HSD and CYP19A as well as the estrogen receptor ESR1 and ESR2. We found the pcDNA3.1-STAT4 markedly decreased the mRNA expressions of CYP17 (Fig. 5a), 3B-HSD (Fig. 5a), 17B-HSD (Fig. 5a), CYP19A (Fig. 5a), ESR1 (Fig. 5e), and ESR2 (Fig. 5e), and decreased concentration of E2 in porcine GCs (Fig. 5c). Whereas the STAT4-siRNA significantly upregulated the mRNA expressions of Star and CYP19A (Fig. 5b), up-regulated the mRNA levels of ESR1 and ESR2 (Fig. 5f) as well as the concentration of E2 in porcine GCs (Fig. 5d). These observations suggested that the STAT4 might suppress the synthesis of E2 in ovarian GCs. Moreover, the STAT4-siRNA could significantly decrease the cell apoptosis rate of pcDNA3.1-KISS1 and significantly increase the concentration of E2 of pcDNA3.1-KISS1, compared to that of the pcDNA3.1-STAT4 (Fig. 6a,b). Moreover, the mRNA level of STAT4 was observed to significantly decrease from immature to mature follicles (Fig. 6c), which was inversed with that of KISS1 during the follicular maturation [6]. These results indicated that the STAT4 might repress the anti-apoptotic effect and the E2 synthetic capacity of KISS1 in GCs to inhibit the follicular development in pigs.
Conclusions
Taken together, the STAT4 might directly target at − 305/− 295 region of KISS1 to negatively regulate the transcription and biological of KISS1, involve in PI3K signaling pathway to promote the cell apoptosis, and participate in estrogen synthesis signaling pathway to suppress the synthesis of E2 in GCs to arrest the follicular development. These proposed works would contribute to providing new biological insight for further investigation on STAT4 and KISS1 in the follicular development of mammals.
Acknowledgements
Not applicable.
Abbreviations
- GCs
Granulosa cells
- KISS1
Kiss-1 metastasis-suppressor gene
- STAT
Signal transducer and activator of transcription
- E2
Estradiol-17β
- PI3K
Phosphatidylinositol 3-OH-kinase
- SiRNA
Small interfering RNA
- ChIP
Chromatin immunoprecipitation
- PBS
Phosphate-buffered saline
- SD
Standard deviation
Authors’ contributions
Conceptualization, Xiaolong Yuan, Xiaoping Xin and Jiaqi Li; Data curation, Xiaolong Yuan; Formal analysis, Xiaolong Yuan; Investigation, Xiaolong Yuan, Xiaoping Xin, Ailing Zhang and Jiaqi Li; Methodology, Xiaoping Xin, Xiangchun Pan and Yao Jiang; Supervision, Zhe Zhang; Validation, Hao Zhang; Writing – original draft, Yao Jiang, Xiaoping Xin and Xiaolong Yuan; Writing – review & editing, Ailing Zhang and Jiaqi Li. The authors read and approved the final manuscript.
Funding
This research was funded by National Key R&D Program of China (2018YFD0501200), the Youth Innovative fund of Guangdong Education Department (2018KQNCX019), the Special Fund for Science and Technology Innovation of Guangdong Province (2018B020203003), the Natural Science Foundation of Guangdong Province (2019A1515010676), and the earmarked fund for China Agriculture Research System (CARS-35).
Availability of data and materials
The dataset supporting the conclusions of this article are included within the article.
Ethics approval and consent to participate
All experiments in the present study were performed in accordance with the guidelines of the Animal Care and Use Committee of South China Agricultural University Guangzhou, China (Approval Number: SCAU#2013–10).
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yao Jiang, Email: jyao192@163.com.
Xiaoping Xin, Email: xiaopingxin1991@163.com.
Xiangchun Pan, Email: 15186255977@126.com.
Ailing Zhang, Email: zhangailing@gdei.edu.cn.
Zhe Zhang, Email: zhezhang@scau.edu.cn.
Jiaqi Li, Email: jqli@scau.edu.cn.
Xiaolong Yuan, Email: yxl@scau.du.cn.
References
- 1.Lomniczi A, Wright H, Ojeda SR. Epigenetic regulation of female puberty. Front Neuroendocrinol. 2015;36:90–107. doi: 10.1016/j.yfrne.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. Nat Neurosci. 2013;16(3):281–289. doi: 10.1038/nn.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourguignon JP, Juul A. Normal female puberty in a developmental perspective. Endocr Dev. 2012;22:11–23. doi: 10.1159/000326624. [DOI] [PubMed] [Google Scholar]
- 4.Zeydabadi Nejad S, Ramezani Tehrani F, Zadeh-Vakili A. The role of Kisspeptin in female reproduction. Int J Endocrinol Metab. 2017;15(3):e44337. doi: 10.5812/ijem.44337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasco V, Pinto FM, Fernandez-Atucha A, Prados N, Tena-Sempere M, Fernandez-Sanchez M, Candenas L. Altered expression of the kisspeptin/KISS1R and neurokinin B/NK3R systems in mural granulosa and cumulus cells of patients with polycystic ovarian syndrome. J Assist Reprod Genet. 2019;36(1):113–120. doi: 10.1007/s10815-018-1338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xin X, Li Z, Zhong Y, Li Q, Wang J, Zhang H, Yuan X, Li J, Zhang Z. KISS1 Suppresses Apoptosis and Stimulates the Synthesis of E2 in Porcine Ovarian Granulosa Cells. Animals (Basel). 2019;9(2):54. [DOI] [PMC free article] [PubMed]
- 7.Basini G, Grasselli F, Bussolati S, Ciccimarra R, Maranesi M, Bufalari A, Parillo F, Zerani M. Presence and function of kisspeptin/KISS1R system in swine ovarian follicles. Theriogenology. 2018;115:1–8. doi: 10.1016/j.theriogenology.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Castellano JM, Gaytan M, Roa J, Vigo E, Navarro VM, Bellido C, Dieguez C, Aguilar E, Sanchez-Criado JE, Pellicer A, et al. Expression of KiSS-1 in rat ovary: putative local regulator of ovulation? Endocrinology. 2006;147(10):4852–4862. doi: 10.1210/en.2006-0117. [DOI] [PubMed] [Google Scholar]
- 9.Shahed A, Young KA. Differential ovarian expression of KiSS-1 and GPR-54 during the estrous cycle and photoperiod induced recrudescence in Siberian hamsters (Phodopus sungorus) Mol Reprod Dev. 2009;76(5):444–452. doi: 10.1002/mrd.20972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cielesh ME, McGrath BM, Scott CJ, Norman ST, Stephen CP. The localization of kisspeptin and kisspeptin receptor in the canine ovary during different stages of the reproductive cycle. Reprod Domest Anim. 2017;52(Suppl 2):24–28. doi: 10.1111/rda.12841. [DOI] [PubMed] [Google Scholar]
- 11.Tanyapanyachon P, Amelkina O, Chatdarong K. The expression of kisspeptin and its receptor in the domestic cat ovary and uterus in different stages of the ovarian cycle. Theriogenology. 2018;117:40–48. doi: 10.1016/j.theriogenology.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Colledge WH. Transgenic mouse models to study Gpr54/kisspeptin physiology. Peptides. 2009;30(1):34–41. doi: 10.1016/j.peptides.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Lapatto R, Pallais JC, Zhang DS, Chan YM, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB. Kiss1(−/−) mice exhibit more variable hypogonadism than Gpr54(−/−) mice. Endocrinology. 2007;148(10):4927–4936. doi: 10.1210/en.2007-0078. [DOI] [PubMed] [Google Scholar]
- 14.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23(48):8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Levy DE. Comparative evolutionary genomics of the STAT family of transcription factors. JAKSTAT. 2012;1(1):23–33. doi: 10.4161/jkst.19418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrian AD, Hatcher MA, Brotman JJ, Galkina EV, Taghavie-Moghadam P, Pei H, Haynes BA, Nadler JL. STAT4 contributes to adipose tissue inflammation and atherosclerosis. J Endocrinol. 2015;227(1):13–24. doi: 10.1530/JOE-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen L, Craig J, Dyce PW, Li J. Cloning of porcine signal transducer and activator of transcription 3 cDNA and its expression in reproductive tissues. Reproduction. 2006;132(3):511–518. doi: 10.1530/rep.1.01055. [DOI] [PubMed] [Google Scholar]
- 18.Hannesdottir L, Daschil N, Philipp S, Tymoszuk P, Muller-Holzner E, Klima G, Verdorfer I, Doppler W. MMTV-neu mice deficient in STAT1 are susceptible to develop ovarian teratomas. Int J Dev Biol. 2012;56(4):279–283. doi: 10.1387/ijdb.113397lh. [DOI] [PubMed] [Google Scholar]
- 19.Hall SE, Upton RMO, McLaughlin EA, Sutherland JM. Phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) follicular signalling is conserved in the mare ovary. Reprod Fert Develop. 2018;30(4):624–633. doi: 10.1071/RD17024. [DOI] [PubMed] [Google Scholar]
- 20.Robker RL, Watson LN, Robertson SA, Dunning KR, McLaughlin EA, Russell DL. Identification of Sites of STAT3 Action in the Female Reproductive Tract through Conditional Gene Deletion. Plos One. 2014;9(7):e101182. [DOI] [PMC free article] [PubMed]
- 21.Gasperin BG, Rovani MT, Ferreira R, Ilha GF, Bordignon V, Goncalves PB, Duggavathi R. Functional status of STAT3 and MAPK3/1 signaling pathways in granulosa cells during bovine follicular deviation. Theriogenology. 2015;83(3):353–359. doi: 10.1016/j.theriogenology.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Ilha GF, Rovani MT, Gasperin BG, Antoniazzi AQ, Goncalves PB, Bordignon V, Duggavathi R. Lack of FSH support enhances LIF-STAT3 signaling in granulosa cells of atretic follicles in cattle. Reproduction. 2015;150(4):395–403. doi: 10.1530/REP-15-0026. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, Chern BSM, Barton-Smith P, Phoon JWL, Tan TY, Viardot-Foucault V, Ku CW, Tan HH, Chan JKY, Lee YH. Peritoneal fluid cytokines reveal new insights of endometriosis subphenotypes. Int J Mol Sci. 2020;21(10):3515. [DOI] [PMC free article] [PubMed]
- 24.Saatcioglu HD, Cuevas I, Castrillon DH. Control of oocyte reawakening by kit. PLoS Genet. 2016;12(8):e1006215. doi: 10.1371/journal.pgen.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Douville G, Sirard MA. Changes in granulosa cells gene expression associated with growth, plateau and atretic phases in medium bovine follicles. J Ovarian Res. 2014;7:50. doi: 10.1186/1757-2215-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan MI, Dias FC, Dufort I, Misra V, Sirard MA, Singh J. Stable reference genes in granulosa cells of bovine dominant follicles during follicular growth, FSH stimulation and maternal aging. Reprod Fertil Dev. 2016;28(6):795–805. doi: 10.1071/RD14089. [DOI] [PubMed] [Google Scholar]
- 27.Lin P, Rui R. Effects of follicular size and FSH on granulosa cell apoptosis and atresia in porcine antral follicles. Mol Reprod Dev. 2010;77(8):670–678. doi: 10.1002/mrd.21202. [DOI] [PubMed] [Google Scholar]
- 28.Hennet ML, Combelles CM. The antral follicle: a microenvironment for oocyte differentiation. Int J Dev Biol. 2012;56(10–12):819–831. doi: 10.1387/ijdb.120133cc. [DOI] [PubMed] [Google Scholar]
- 29.Almeida CP, Ferreira MCF, Silveira CO, Campos JR, Borges IT, Baeta PG, Silva FHS, Reis FM, Del Puerto HL. Clinical correlation of apoptosis in human granulosa cells-a review. Cell Biol Int. 2018;42(10):1276–1281. doi: 10.1002/cbin.11036. [DOI] [PubMed] [Google Scholar]
- 30.Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 2014;20(1):1–11. doi: 10.1093/humupd/dmt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu CL, Cowan RG, Harman RM, Quirk SM. Cell cycle progression and activation of Akt kinase are required for insulin-like growth factor I-mediated suppression of apoptosis in granulosa cells. Mol Endocrinol. 2004;18(2):326–338. doi: 10.1210/me.2003-0178. [DOI] [PubMed] [Google Scholar]
- 32.Zheng W, Nagaraju G, Liu Z, Liu K. Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Mol Cell Endocrinol. 2012;356(1–2):24–30. doi: 10.1016/j.mce.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Li Q, He H, Zhang YL, Li XM, Guo X, Huo R, Bi Y, Li J, Fan HY, Sha J. Phosphoinositide 3-kinase p110delta mediates estrogen- and FSH-stimulated ovarian follicle growth. Mol Endocrinol. 2013;27(9):1468–1482. doi: 10.1210/me.2013-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Sheng B, Zhao M, Shen Q, Zhu H, Zhu X. The prognostic values of signal transducers activators of transcription family in ovarian cancer. Biosci Rep. 2017;37(4):BSR20170650. [DOI] [PMC free article] [PubMed]
- 35.Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta 2. Immunity. 2012;37(3):501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ieda N, Uenoyama Y, Tajima Y, Nakata T, Kano M, Naniwa Y, Watanabe Y, Minabe S, Tomikawa J, Inoue N, et al. KISS1 gene expression in the developing brain of female pigs in pre- and peripubertal periods. J Reprod Dev. 2014;60(4):312–316. doi: 10.1262/jrd.2013-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuo Y, Zhou D, Che L, Fang Z, Lin Y, Wu D. Feeding prepubescent gilts a high-fat diet induces molecular changes in the hypothalamus-pituitary-gonadal axis and predicts early timing of puberty. Nutrition. 2014;30(7–8):890–896. doi: 10.1016/j.nut.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Tsunoda T, Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15(7–8):622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- 39.Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan X, Zhou X, He Y, Zhong Y, Zhang A, Zhang Z, Zhang H, Li J. C/EBPbeta promotes STAT3 expression and affects cell apoptosis and proliferation in porcine ovarian granulosa cells. Genes (Basel). 2018;9(6):295. [DOI] [PMC free article] [PubMed]
- 41.Yuan X, Deng X, Zhou X, Zhang A, Xing Y, Zhang Z, Zhang H, Li J. MiR-126-3p promotes the cell proliferation and inhibits the cell apoptosis by targeting TSC1 in the porcine granulosa cells. In Vitro Cell Dev Biol Anim. 2018;54(10):715–724. doi: 10.1007/s11626-018-0292-0. [DOI] [PubMed] [Google Scholar]
- 42.Khristi V, Chakravarthi VP, Singh P, Ghosh S, Pramanik A, Ratri A, Borosha S, Roby KF, Wolfe MW, Rumi MAK. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol Cell Endocrinol. 2018;474:214–26. [DOI] [PubMed]
- 43.de Assis Rodrigues NP, Lagana AS, Zaia V, Vitagliano A, Barbosa CP, de Oliveira R, Trevisan CM, Montagna E. The role of Kisspeptin levels in polycystic ovary syndrome: a systematic review and meta-analysis. Arch Gynecol Obstet. 2019;300(5):1423–1434. doi: 10.1007/s00404-019-05307-5. [DOI] [PubMed] [Google Scholar]
- 44.Schams D, Berisha B. Steroids as local regulators of ovarian activity in domestic animals. Domest Anim Endocrin. 2002;23(1–2):53–65. doi: 10.1016/S0739-7240(02)00145-5. [DOI] [PubMed] [Google Scholar]
- 45.Conley AJ, Howard HJ, Slanger WD, Ford JJ. Steroidogenesis in the preovulatory porcine follicle. Biol Reprod. 1994;51(4):655–661. doi: 10.1095/biolreprod51.4.655. [DOI] [PubMed] [Google Scholar]
- 46.Matsuda F, Inoue N, Manabe N, Ohkura S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. 2012;58(1):44–50. doi: 10.1262/jrd.2011-012. [DOI] [PubMed] [Google Scholar]
- 47.Valdez KE, Turzillo AM. Regulation of nuclear factor-kappaB (NF-kappaB) activity and apoptosis by estradiol in bovine granulosa cells. Mol Cell Endocrinol. 2005;243(1–2):66–73. doi: 10.1016/j.mce.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Chou CH, Chen MJ. The effect of steroid hormones on ovarian follicle development. Vitam Horm. 2018;107:155–175. doi: 10.1016/bs.vh.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Depalo R, Cavallini A, Lorusso F, Bassi E, Totaro I, Marzullo A, Bettocchi S, Selvaggi L. Apoptosis in normal ovaries of women with and without endometriosis. Reprod BioMed Online. 2009;19(6):808–815. doi: 10.1016/j.rbmo.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 50.Vetvicka V, Lagana AS, Salmeri FM, Triolo O, Palmara VI, Vitale SG, Sofo V, Kralickova M. Regulation of apoptotic pathways during endometriosis: from the molecular basis to the future perspectives. Arch Gynecol Obstet. 2016;294(5):897–904. doi: 10.1007/s00404-016-4195-6. [DOI] [PubMed] [Google Scholar]
- 51.Kralickova M, Lagana AS, Ghezzi F, Vetvicka V. Endometriosis and risk of ovarian cancer: what do we know? Arch Gynecol Obstet. 2020;301(1):1–10. doi: 10.1007/s00404-019-05358-8. [DOI] [PubMed] [Google Scholar]
- 52.Reddy P, Adhikari D, Zheng W, Liang S, Hamalainen T, Tohonen V, Ogawa W, Noda T, Volarevic S, Huhtaniemi I, et al. PDK1 signaling in oocytes controls reproductive aging and lifespan by manipulating the survival of primordial follicles. Hum Mol Genet. 2009;18(15):2813–2824. doi: 10.1093/hmg/ddp217. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L, Huang J, Yang N, Greshock J, Liang S, Hasegawa K, Giannakakis A, Poulos N, O'Brien-Jenkins A, Katsaros D, et al. Integrative genomic analysis of phosphatidylinositol 3′-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clin Cancer Res. 2007;13(18 Pt 1):5314–5321. doi: 10.1158/1078-0432.CCR-06-2660. [DOI] [PubMed] [Google Scholar]
- 54.Adhikari D, Flohr G, Gorre N, Shen Y, Yang HR, Lundin E, Lan ZJ, Gambello MJ, Liu K. Disruption of Tsc2 in oocytes leads to overactivation of the entire pool of primordial follicles. Mol Hum Reprod. 2009;15(12):765–770. doi: 10.1093/molehr/gap092. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Castrillon DH, Zhou W, Richards JS. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol Endocrinol. 2013;27(2):238–252. doi: 10.1210/me.2012-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, Zhang L, Wang B, Wei R, Wang Y, Wan J, Zhang C, Zhao L, Zhu X, Zhang Y, et al. MiR-337-3p suppresses proliferation of epithelial ovarian cancer by targeting PIK3CA and PIK3CB. Cancer Lett. 2020;469:54–67. doi: 10.1016/j.canlet.2019.10.021. [DOI] [PubMed] [Google Scholar]
- 57.Cao XH, Wang XY, Lu LL, Li XY, Di R, He XY, Hu WP, Zeng XY, Liu QY, Chu MX. Expression and functional analysis of the BCL2-associated agonist of cell death (BAD) gene in the sheep ovary during the reproductive cycle. Front Endocrinol. 2018;19(9):512. [DOI] [PMC free article] [PubMed]
- 58.Lv L, Zhang J, Huang X, Zhao Y, Zhou Z, Zhang H. Lentivirus-mediated RNA interference targeting STAT4 inhibits the proliferation of vascular smooth muscle cells. Arch Med Res. 2008;39(6):582–589. doi: 10.1016/j.arcmed.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 59.Hasegawa T, Kamada Y, Hosoya T, Fujita S, Nishiyama Y, Iwata N, Hiramatsu Y, Otsuka F. A regulatory role of androgen in ovarian steroidogenesis by rat granulosa cells. J Steroid Biochem Mol Biol. 2017;172:160–165. doi: 10.1016/j.jsbmb.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 60.Owens LA, Abbara A, Lerner A, O'Floinn S, Christopoulos G, Khanjani S, Islam R, Hardy K, Hanyaloglu AC, Lavery SA, et al. The direct and indirect effects of kisspeptin-54 on granulosa lutein cell function. Hum Reprod. 2018;33(2):292–302. doi: 10.1093/humrep/dex357. [DOI] [PubMed] [Google Scholar]
- 61.Garg D, Merhi Z. Relationship between advanced glycation end products and steroidogenesis in PCOS. Reprod Biol Endocrin. 2016;14(1):71. [DOI] [PMC free article] [PubMed]
- 62.Michalovic L, Currin L, Gutierrez K, Bellefleur AM, Glanzner WG, Schuermann Y, de Macedo MP, Bohrer RC, Dicks N, Lopez R, et al. Granulosa cells of prepubertal cattle respond to gonadotropin signaling and upregulate genes that promote follicular growth and prevent cell apoptosis. Mol Reprod Dev. 2018;85(12):909–920. doi: 10.1002/mrd.23066. [DOI] [PubMed] [Google Scholar]
- 63.Song D, Huang XL, Hong L, Yu JM, Zhang ZF, Zhang HQ, Sun ZG, Du J. Sequence variants in FSHR and CYP19A1 genes and the ovarian response to controlled ovarian stimulation. Fertil Steril. 2019;112(4):749. doi: 10.1016/j.fertnstert.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Song D, Huang XL, Hong L, Yu JM, Zhang ZF, Zhang HQ, Sun ZG, Du J. Sequence variants in FSHR and CYP19A1 genes and the ovarian response to controlled ovarian stimulation. Fertil Steril. 2019;112(4):749–57. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article are included within the article.