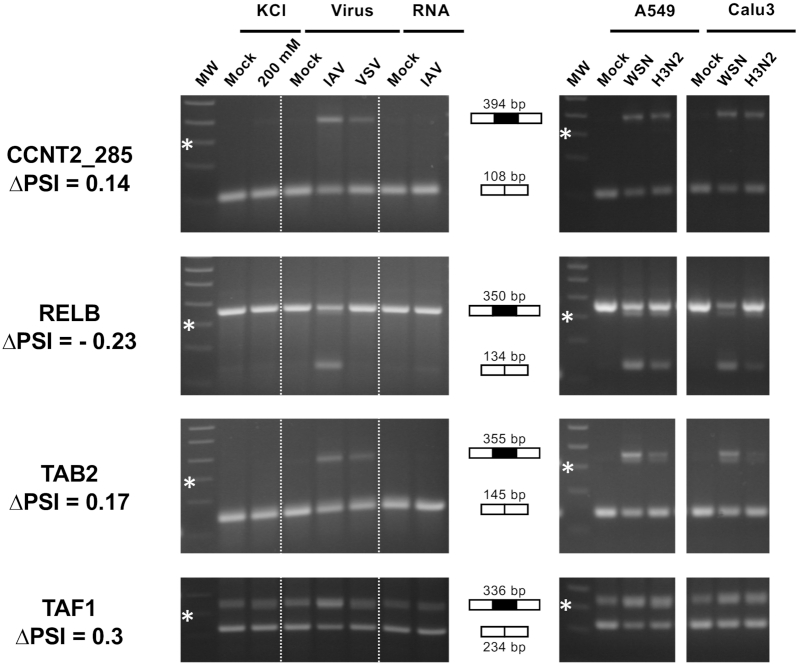
Figure 3.
RT-PCR validation and further characterization of IAV-sensitive splicing events detected by RNA-seq. (Left) A549 cells were subjected to viral infection at a high MOI (WSN versus VSV or mock infection), RNA transfection (RNA extracted from WSN-infected versus mock-infected cells) or osmotic stress (200 mM KCl versus mock treatment). (Right) A549 or Calu-3 cells were infected at a high MOI with the WSN virus or a seasonal H3N2 IAV, or mock infected. Total RNA was extracted and RT-PCR was performed using primers flanking the exon of interest. The amplification products were loaded on a 2% agarose gel. The expected size in case of exon inclusion or exclusion is indicated, as well as the ΔPSI value as determined from RNA-seq data. MW: molecular weight. The 300-bp band of the 100-bp DNA Ladder (NEB) is indicated by a star.