Figure 1.
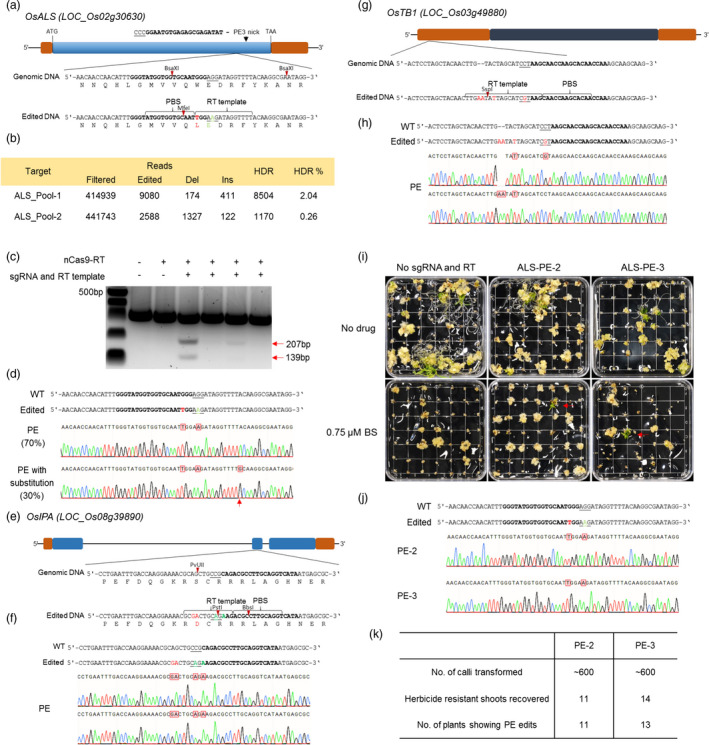
Prime editing of OsALS for herbicide resistance. (a) Schematic representation of rice ALS locus. A single nucleotide substitution TGG to TTG (W548L) produced a herbicide resistance in rice. The repair template (RT) designed with two substitutions, G to T for herbicide resistance and a silent mutation G to A to destroy PAM site. These substitutions generate MfeI site and abolish the BsaXI site. The exon is indicated as blue box. (b) The rice callus was transformed with ALS‐PE2 (nCas9‐RT_pegRNA) via agrobacterium. After two rounds of selections of T‐DNA on hygromycin, the proliferating rice calli were pooled and used for amplicon deep sequencing. (c) The amplicons from different rice calli were enriched for editing by BsaXI and after purification, PCR was done and amplicons were digested with MfeI. The digested production indicates the editing in the cells and further confirmed via Sanger sequencing (d). Some of the edited reads, indicated by arrow, were also showing A to G substitutions. This G probably corresponds to the first base of the scaffold RNA adjacent to RT template in pegRNA. (e) Schematic of rice locus IPA (Ideal Plant Architecture). We have designed a pegRNA for two consecutive substitutions AG to GA to convert S163 to D. Two silent substitutions have been done CGC to AGA which convert R165 to R and destroy the PAM site. By these mutations, PvUII site was lost and two sites Pst1 and BbsI were generated. (f) The PvUII enriched DNA samples are confirmed by Sanger sequencing. (g) Schematic of rice locus TB1 (TEOSINTE BRANCHED1). We have designed a pegRNA to target the GTAC motif in promoter of OsTB1. In the repair, template C was converted to G to destroy PAM site. Two consecutive insertions AA and one substitution were done to destroy binding motif. These mutations also created SspI restriction site and destroy RsaI site. (h) The RsaI enriched DNA samples are confirmed by Sanger sequencing. (i) The ALS‐PE‐2 and ALS‐PE‐3 plasmids were transformed in rice via agrobacterium. After selection, the regeneration was done with 0.75 µm BS. Arrows indicate the regeneration of herbicide‐resistant shoots. (j) The selected PCR fragments were analysed by Sanger sequencing. (k) The number of plants recovered from PE‐2 and PE‐3 is almost equal. Some of the herbicide resistance plants are homozygous.
