Figure 1.
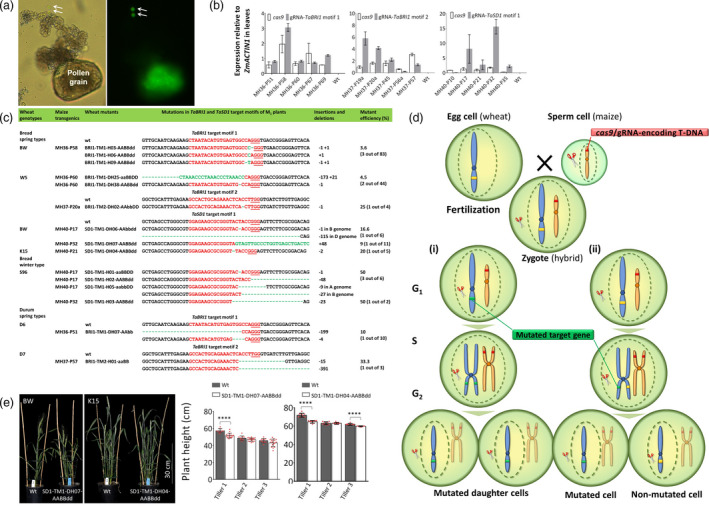
Site‐directed mutagenesis in bread and durum wheat via pollination by cas9/gRNA‐transgenic maize. (a) GFP accumulation in maize sperm cells (arrows); bright field (left) and GFP (right) filter images with 200x magnification. (b) Expression of cas9 and gRNAs. Graphs represent the mean values of cas9 and gRNA, and error bars represent the standard deviation derived from three replications. (c) The TaBRI1 and TaSD1 target motifs are highlighted in red with the protospacer‐adjacent motifs (PAM) being underlined. Mutations are indicated by green font, and the numbers given to the right represent the concerned nucleobases. Mutant efficiency is the proportion of plants with mutations out of the total number of plants analysed. Abbreviations: (H/DH) haploid/doubled haploid, (TM) target motif, (wt) wild type, (+) insertions and (−) deletions. (d) Mutations induced in hybrid zygotes at various phases before and after mitosis. (e) Tiller height of representative doubled‐haploid SD1 M2 plants at anthesis stage. The graph represents the mean height values of the first three tillers from the BW‐wt (n = 10), SD1‐TM1‐DH07‐AABBdd mutant (n = 17), K15‐wt (n = 10) and SD1‐TM1‐DH04‐AABBdd mutant (n = 10). Significant differences between mutants and wt counterparts are indicated by asterisks, with **** representing a P‐value < 0.0001 according to unpaired t‐test; n is the number of plants analysed, and error bars represent the standard deviation.
