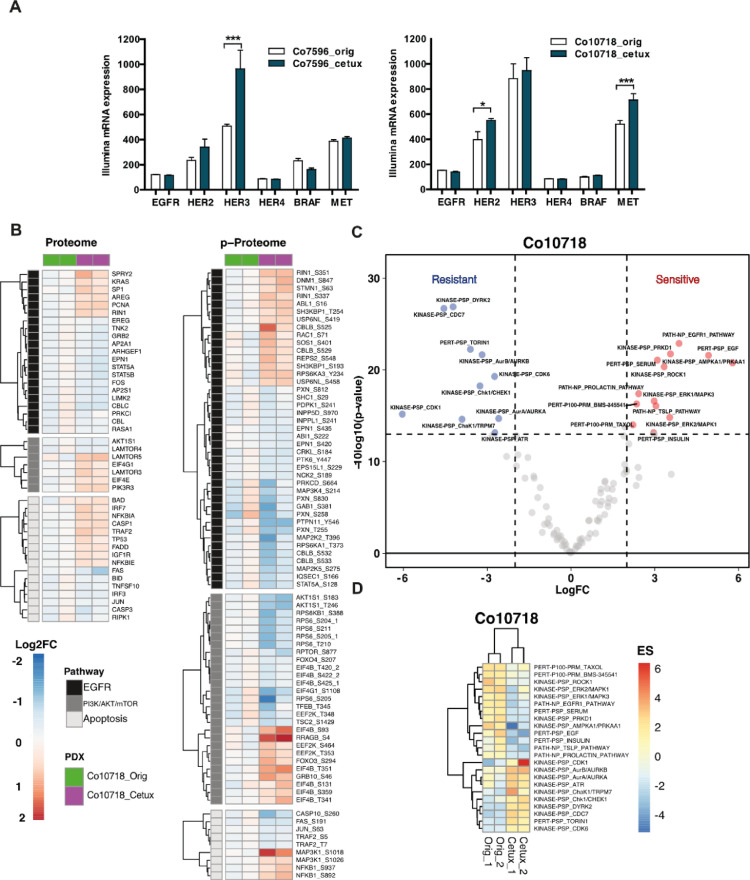
Figure 6.
Gene expression and proteome, phosphoproteome analysis of cetuximab sensitive vs resistant PDX. (A) Comparison of the gene expression of EGFR receptors and molecules involved in EGFR signal transduction. (B) Heatmaps of significantly changed proteins and phosphosites (One-sample t test, adj. P value < 0.1, reproducibility filter = 0.01) within the EGFR, PI3K/AKT/Mtor, and apoptosis pathways (Wiki Pathway annotations) for the Co10718 model. P values are calculated with data of 2 replicates for the Co10718_cetux models normalized against Co10718_orig models. The annotation column shows 2 lanes for 2 replicates of sensitive (Co10718_orig) and cetuximab resistant (Co10718_cetux) models with green and magenta colors, respectively. On the heatmap, blue color indicates down-regulation whereas red color corresponds to proteins and phosphosites up-regulated in the cetuximab resistant comparing to sensitive pair. (C) Volcano plot of the P values vs the logFC (fold change) of enrichment scores of PTM-SEA (Ref DOI: 10.1074/mcp.TIR118.000943). Two-sample t test is performed between resistant and sensitive replicates of the Co10718 model. Proteins crossing the significance lines (|logFC| ≥ 2, Pvalue ≤ 0.05) are colored in blue or red. (D) Heatmap depicting enrichment scores of significantly regulated pathways in the PTM-SEA analysis.