Figure 1.
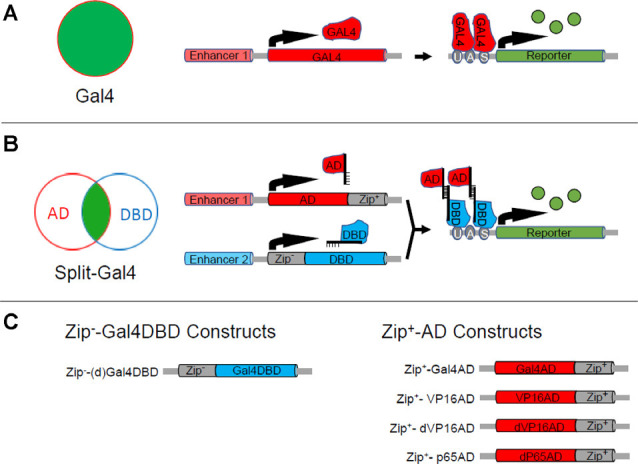
The Split Gal4 system. (A) The binary Gal4-UAS expression system can be used to target the expression of a reporter or effector (green) to a group of cells (red circle) in which an enhancer (Enhancer 1) is active. As illustrated in the right-hand schematics, Enhancer 1 drives expression of the Gal4 transcription factor (red) which in turn drives expression of the reporter or effector gene (green), which is placed downstream of Gal4’s DNA binding site (UAS). (B) The Split Gal4 system uses two enhancers with activity in overlapping cell groups (red and blue circles) to target reporter or effector expression (green) to the intersection of the two groups. The intersectional logic of expression is shown schematically in the Venn diagram (left). The right-hand panels illustrate the transcriptional mechanisms: one enhancer (Enhancer 1) is used to target expression of the transcription activation domain (AD) of Gal4 or some other transcription factor fused to the Zip+ leucine zipper, while the other (Enhancer 2) is used to target expression of the Gal4 DNA binding domain (Gal4DBD) fused to the Zip+ leucine zipper. Association of Zip+ and Zip+ brings the GalDBD and AD components together to reconstitute Gal4 transcriptional activity and drive expression of UAS-transgenes. (C) Design of Zip−-Gal4DBD and Zip+-AD constructs. Two Zip−-Gal4DBD constructs have been made. They share the same sequence, but the one made by Pfeiffer et al. (2010; designated here as dGal4DBD) is codon-optimized for use in Drosophila and is placed behind a Drosophila synthetic core promoter. Activation domains from three different transcription factors have been used to make Zip+-AD constructs. Zip+-p65AD and Zip+-dVP16AD are codon-optimized and show strong, high-fidelity expression.
