Figure 4.
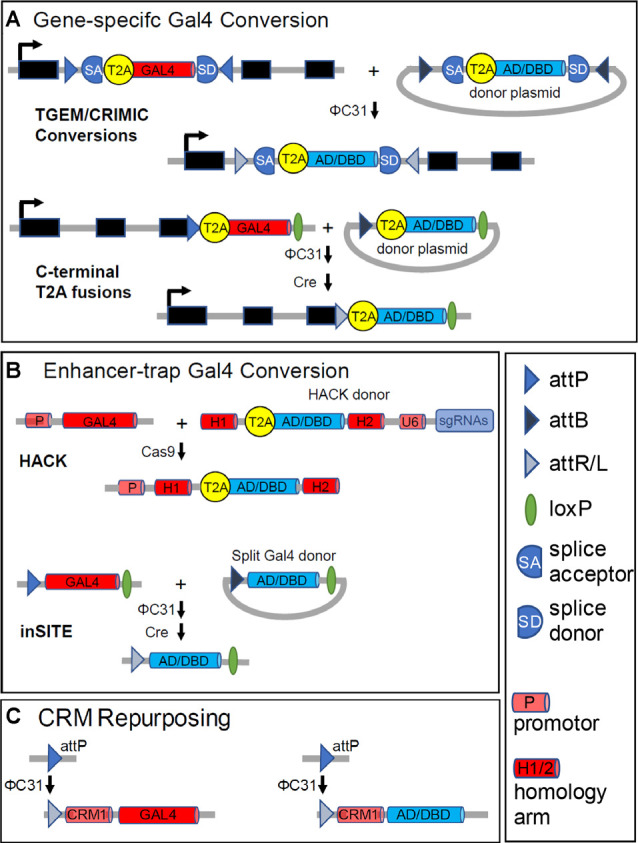
Generating Split Gal4 hemidrivers from Gal4 drivers. While Split Gal4 hemidriver lines can be generated directly, in most instances a Gal4 driver with a desired pattern of expression serves as an intermediate. These starting Gal4 drivers fall into three classes: (A) “gene-trap” Gal4 lines with gene-specific expression; (B) enhancer-trap Gal4 lines made by transposon-mediated transformation; and (C) CRM-Gal4 lines that use enhancer-containing cis-regulatory modules with defined sequences. (A) Gene-trap drivers typically couple Gal4 expression to that of a native gene using T2A peptides (see text). T2A-Gal4 constructs are inserted either intronically, as in TGEM or CRIMIC lines (top), or just before the stop codon in the coding sequence (bottom). In both cases, the inclusion of either attP sites or an attP and a loxP site flanking the inserted constructs permits conversion of the Gal4 line into a Split Gal4 hemidriver. A Zip+-AD or Zip−-Gal4DBD donor construct with complementary flanking attB sites (or an attB and loxP site) can be substituted for Gal4, as indicated. (B) Top panels: the CRISPR/Cas-based HACK method (Lin and Potter, 2016; Xie et al., 2018) can be used to convert arbitrary enhancer-trap Gal4 drivers into Split Gal4 hemidrivers using a universal donor construct. The HACK donor construct has a T2A- Zip+-AD or -Zip−-DNA-BINDING (DBD) sequence flanked by homology arms (H1 and H2) taken from the Gal4 coding sequence. Also, the donor construct has an expression module for guide RNAs targeted to sites in the Gal4 sequence separating H1 and H2. Cas9-mediated cleavage of Gal4 at these sites, followed by homology-directed repair inserts the desired T2A-Split Gal4 construct in-frame into the—now broken—Gal4 sequence. Bottom panels: Enhancer-trap Gal4 lines made using the inSITE system (Gohl et al., 2011) can be converted into Split Gal4 hemidrivers by a series of genetic crosses. The system uses a set of three recombinases (Flp is omitted from the figure for simplicity) to substitute the desired Split Gal4 construct for Gal4 at the site of insertion. (C) The sparsely-expressing Gal4 lines made by the Rubin and Dickson labs (Jenett et al., 2012; Kvon et al., 2014) use enhancer fragments with defined sequences (CRMs) to drive Gal4 expression in specific patterns. The CRM-Gal4 constructs are also inserted into defined attP landing sites. A Split Gal4 hemidriver corresponding to a given CRM-Gal4 driver can thus be made by inserting into the identical genomic site (e.g., attP) a construct that uses the same CRM (here “CRM1”) to drive a Zip+-p65AD or Zip−-Gal4DBD construct instead of Gal4. As Dionne et al. (2018, #114) caution, insertion of the Split Gal4 construct into other genomic sites may lead to deviations from the original expression pattern. The legend indicates symbols used for various DNA motifs.
