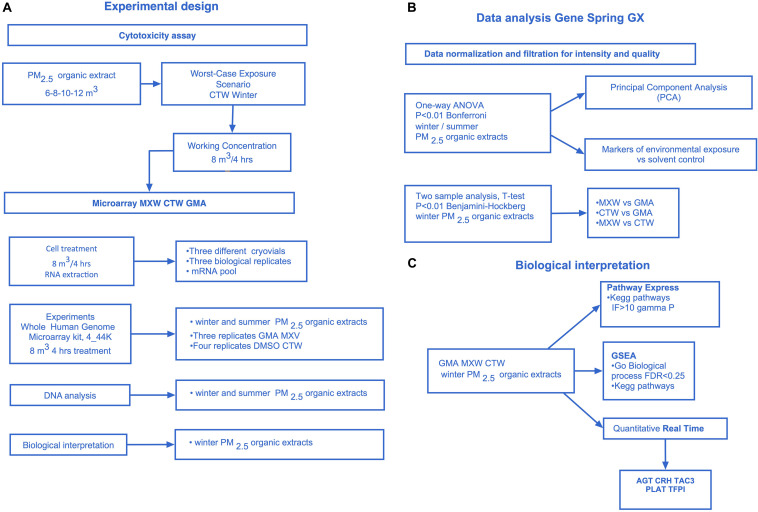
FIGURE 1.
Experimental design. (A) The experimental design. T47D Cytotoxicity assay, Microarray experiments (whole human genome, 4 × 44k): winter and summer PM2.5 extracts 8 m3 4 h treatment, Data analysis: winter and/or summer PM2.5 extracts, Biological interpretation: winter PM2.5 extracts. (B) GeneSpring (GeneSpring GX, Agilent Technologies) Data analysis. Data normalization and filtration for intensity and quality signal, One-Way ANOVA Analysis (winter and summer PM2.5 extracts, P < 0.01 Bonferroni, 11,483 differentially expressed genes) to select differentially expressed genes among the treatments and the control, Principal Component Analysis (PCA), Analysis of markers of environmental exposure (CYP1A1, CYP1B1, HMOX1), t-test analysis on winter PM2.5 samples: MXW vs. GMA, CTW vs. GMA, MXW vs. CTW (P < 0.01 Benjamini–Hockberg). (C) Biological interpretation on winter PM2.5 extracts. Microarray analysis: Pathway express (PE), Gene Set Enrichment Analysis (GSEA). Real time PCR: AGT, CRH, TAC3, PLAT, TFPI.