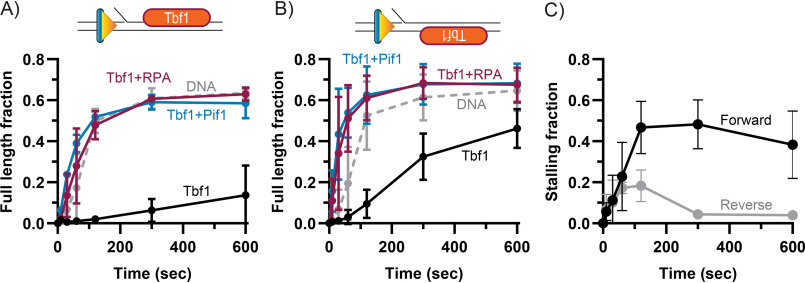
Figure 2.
RPA is sufficient to stimulate DNA synthesis through a Tbf1 block. A, quantification of full-length product formation during primer extension assays performed using Pol δDV with and without Tbf1, RPA, and Pif1. B, same as A, but for a DNA substrate with the Tbf1 binding logo on the opposite strand (reverse substrate). C, quantification of the fraction of stalling products (forward: +13-16 nt; reverse: +7-10) formed by Pol δDV in the absence of RPA and with Tbf1 binding logo either in the forward or reverse orientations. In A–C, the error bars are the mean ± S.D. from 3 independent replicates.